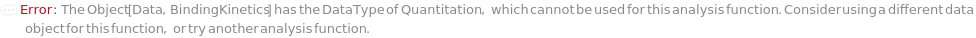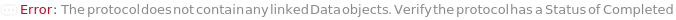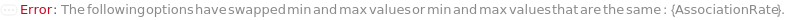AnalyzeBindingKinetics
AnalyzeBindingKinetics[AssociationData, DissociationData]⟹Object
solves for kinetic association and dissociation rates such that the model described by 'reactions' fits the input AssociationData and DissociationData.
AnalyzeBindingKinetics[KineticData]⟹Object
fits to the trajectories in the data objects or protocol from ExperimentBioLayerInterferometry.
Details
- Uses least-squares optimization to solve for the unknown kinetic rates such that the predicted trajectories from the resulting mechanism best fit the training trajectories.
- Global optimization uses NMinimize, while Local optimization uses FindMinimum.
- Simulated trajectories are generated during optimization using SimulateKinetics, which uses NDSolve to numerically solve the differential equations describing the reaction network.
Input

Output

Data Processing Options
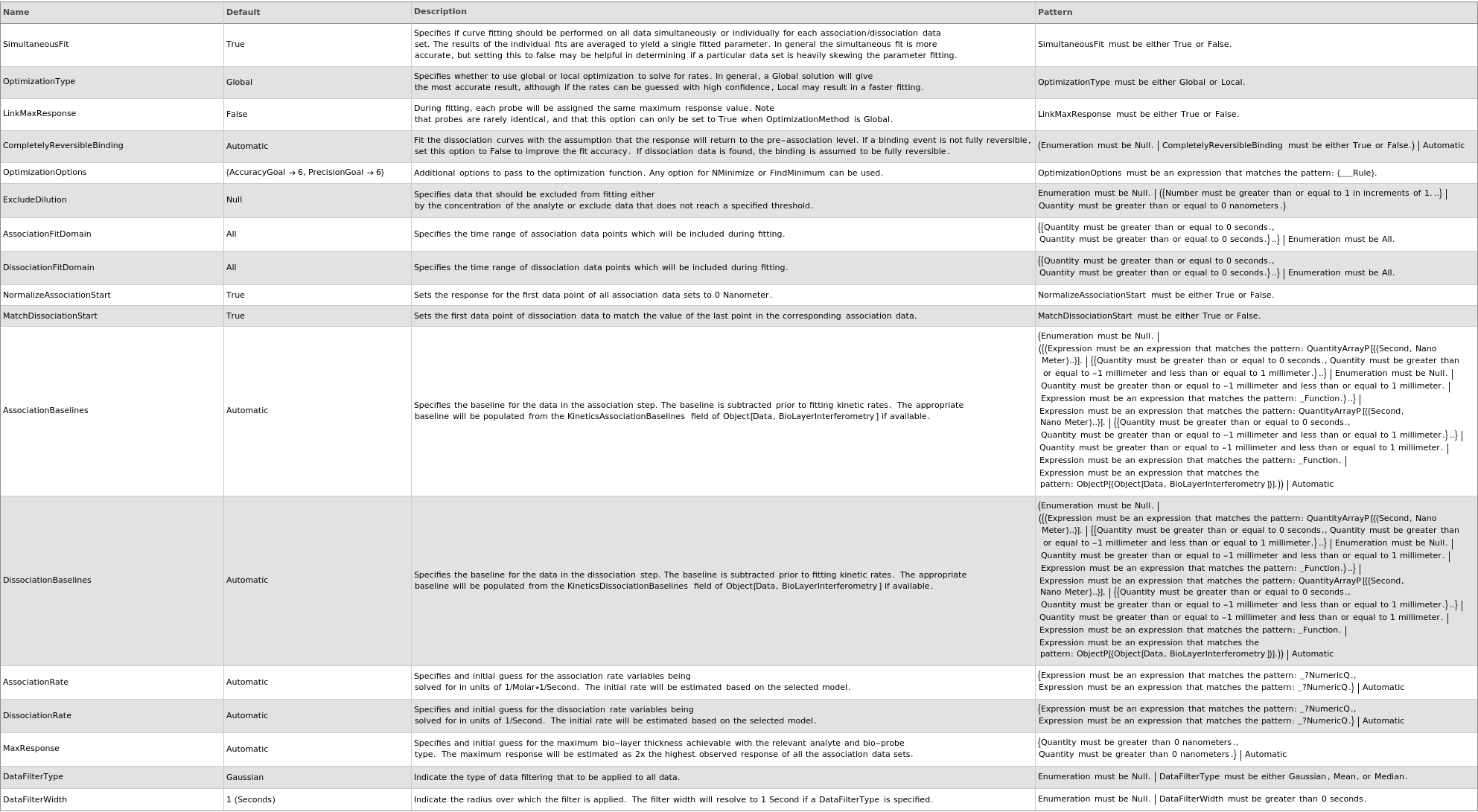
Method Options Options

General Options

Method Options
Examples
open allclose allBasic Examples (3)
AnalyzeBindingKinetics populates the KineticsAnalysis field with a data object containing the relevant fields:
Given an Object[Data,BioLayerInterferometry] with association and dissociation data, AnalyzeBindingKinetics returns an Analysis Object:
Given an Object[Protocol,BioLayerInterferometry] with kinetics Data, AnalyzeBindingKinetics returns an Analysis Object:
Additional Examples (2)
Options (24)
AnalyteDilutions (2)
AssociationBaselines (3)
CompletelyReversibleBinding (1)
DataFilterType (1)
DissociationBaselines (2)
LinkMaxResponse (1)
MatchDissociationStart (1)
NormalizeAssociationStart (1)
Messages (5)
BindingKineticsMultipleAnalytes (1)
InvalidBindingKineticsDataType (1)
NoBindingKineticsData (1)
NoMolecularWeight (1)
Last modified on Wed 17 Sep 2025 17:27:28