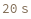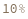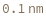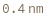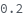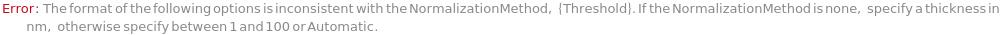AnalyzeEpitopeBinning
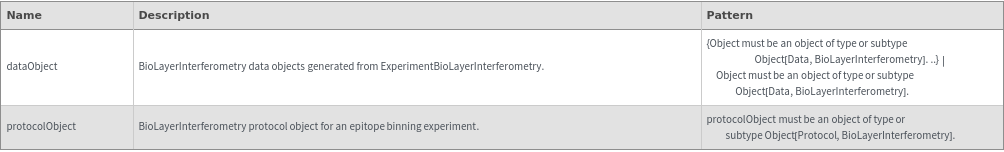
AnalyzeEpitopeBinning[dataObject]⟹object
classifies antibodies by their interaction with a given antigen using dataObject collected by biolayer interferometry.
AnalyzeEpitopeBinning[protocolObject]⟹object
classifies antibodies by their interaction with a given antigen using data found in protocolObject for ExperimentBioLayerInterferometry.
Details
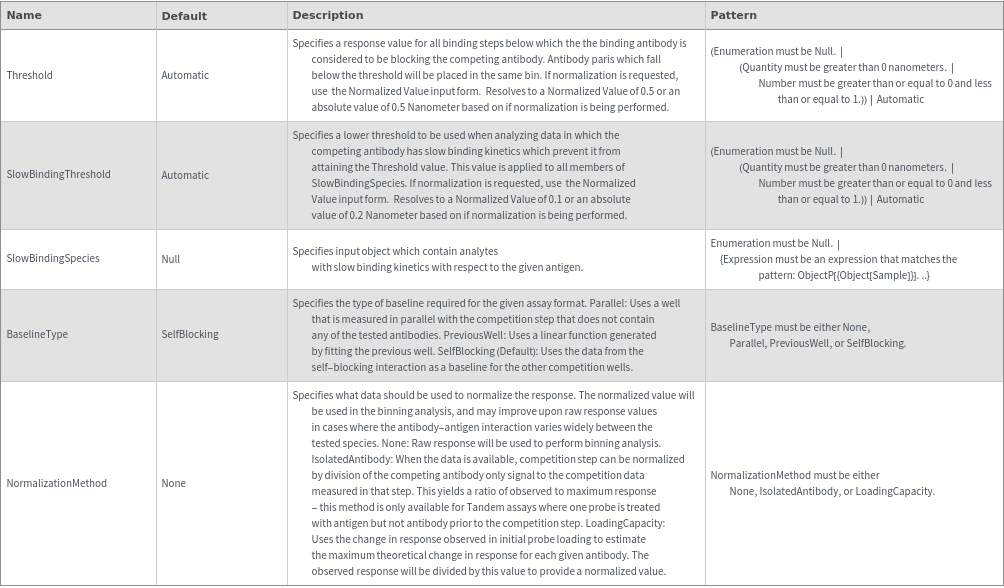
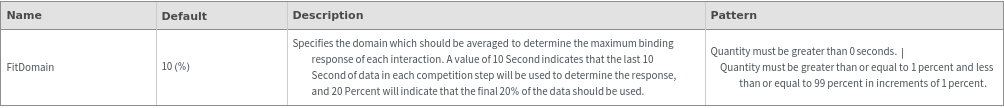
- Cross blocking is quantitated by the maximum bio-layer thickness achieved during the association of the second (competing) antibody. If the change in biolayer thickness during exposure to the secondary (competing) antibody exceeds the specified threshold, the two antibodies are considered to bind distinct epitopes and are placed in different bins. If the change in biolayer thickness does not exceed this threshold, they will be placed in the same bin. There are options to set a low binding threshold for certain species, in the even that some antibody-antigen interactions are sufficiently slow that even with long association times they may not reach a comparable bio-layer thickness to the other measured antibodies.
- The success of this experiment and validity of the analysis is highly dependent on verifying the on and off rates of the tested antibody/antigen pairings. Ensure that the initial antibody/antigen pairing does not have an excessively high dissociation rate, which will invalidate the experimental results.
- All pairings are reported in the A -> B and B -> A binding format.
- Any number of data object can be input to this function, though it is highly recommended that every combination of antibodies is studies for most accurate results. If this is not the case, it is possible that antibodies that bind the same epitope will appear in different bins due to a lack of experimental data.
Input
Output

Data Processing Options
General Options
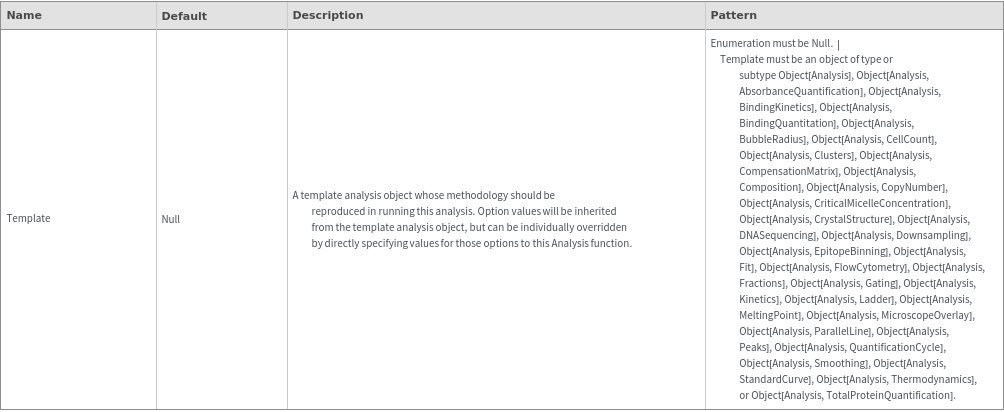
Method Options