ExperimentBioconjugation
ExperimentBioconjugation[SamplePools, NewIdentityModels]⟹Protocol
generates a Protocol object to covalently bind SamplePools through chemical crosslinking to create a sample composed of NewIdentityModels. Bioconjugation reactions are a restricted form of synthesis where conjugations: 1) occur in aqueous solution, 2) occur at atmospheric conditions, 3) are low volume, 4) do not require slow addition of reagents, 5) and do not require reaction monitoring.
Bioconjugation chemically links molecules together where the resulting molecule takes on properties of all parent molecules. This procedure is typically used to modify proteins, antibodies, solid phase supports, and oligomers. Common conjugates are fluorophores, nanoparticles, biotin, and horseradish peroxidase. First, if necessary, the input sample is functionalized or activated to prepare covalent binding sites to accept the conjugate. The activated input sample is then incubated with the conjugates and a covalent bond is formed. The reaction can be quenched to deactivate any unreacted binding sites. Downstream purification may be used to separate conjugated samples from the reaction components prior to use in other assays.
Experimental Principles
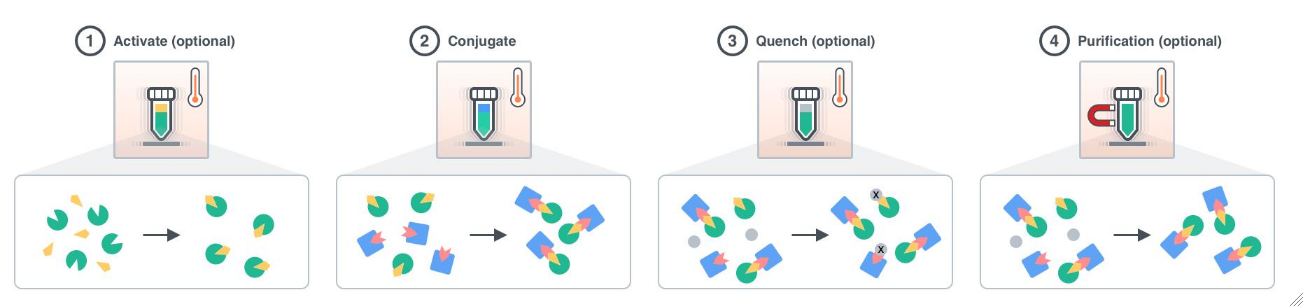
Figure 1.1: Bioconjugation covalently binds a biomolecule to a molecule of interest. Optional Step 1: Biomolecules can be activated to prepare and expose binding sites prior to conjugation. Activation parameters can be used to specify the activation reagent (ActivationReagent, ActivationReagentVolume), mixing of the activation sample (ActivationMix, ActivationMixType, etc.), the incubation time (ActivationTime), the incubation temperature (ActivationTemperature), and any post activation washing if desired (PostActivationWash). Step 2: The conjugation samples are mixed (ConjugationMix, ConjugationMixType, etc) at the desired ratio incubated for the ConjugationTime at the ConjugationTemperature. During this step, covalent bonds are formed between the conjugation samples to create a new molecule characterized by properties of each individual conjugation molecule. Optional Step 3: Unreacted conjugates can be quenched using the QuenchReaction option to deactivate any reactive sites or inhibit the molecular properties of any unconjugated molecules. Step 4: The resulting conjugated molecule can be further purified using PostConjugationWorkup options or used in downstream assays. This process is ideal for small volume conjugations of antibodies, proteins, oligomers, and microbeads including xMap Luminex beads. All steps of this experiment use ExperimentSampleManipulation primitives such as Transfer, Incubate, Filter, and Pellet. For more information on options available in these primitives or to understand how ExperimentSampleManipulation works please see the corresponding help file.
Instrumentation

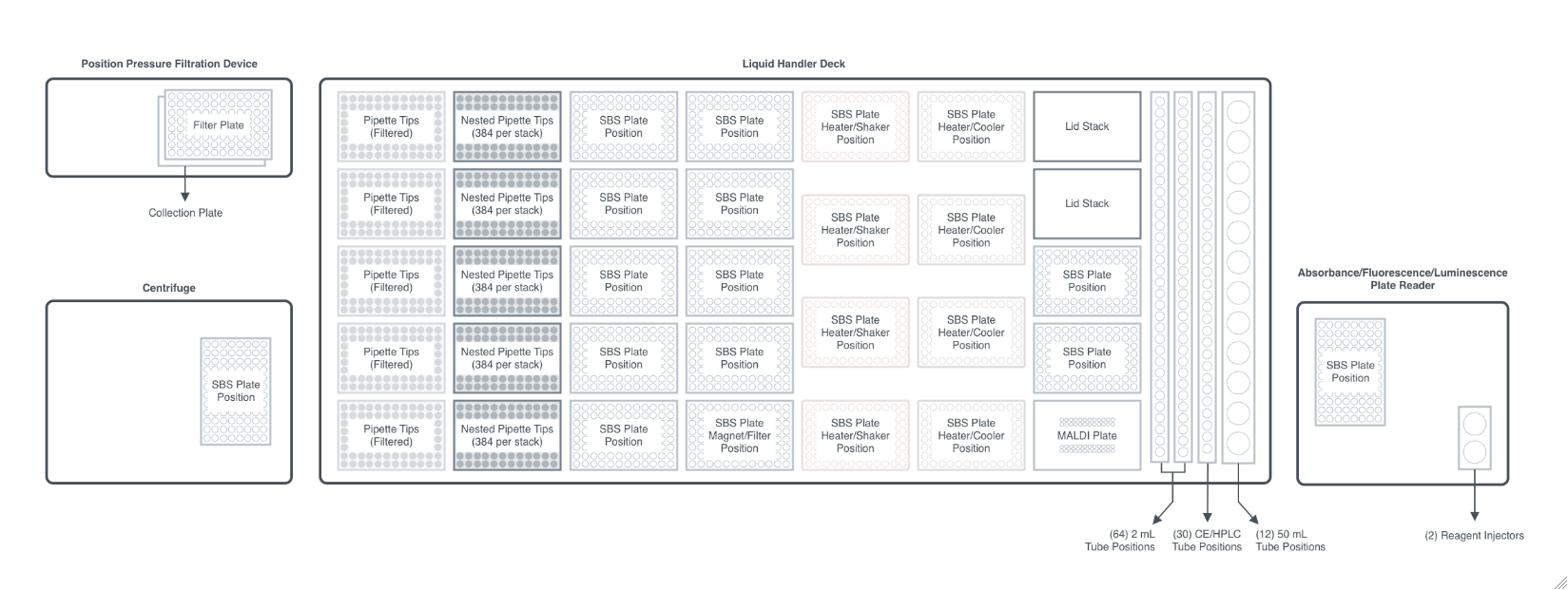
Hamilton STARlet
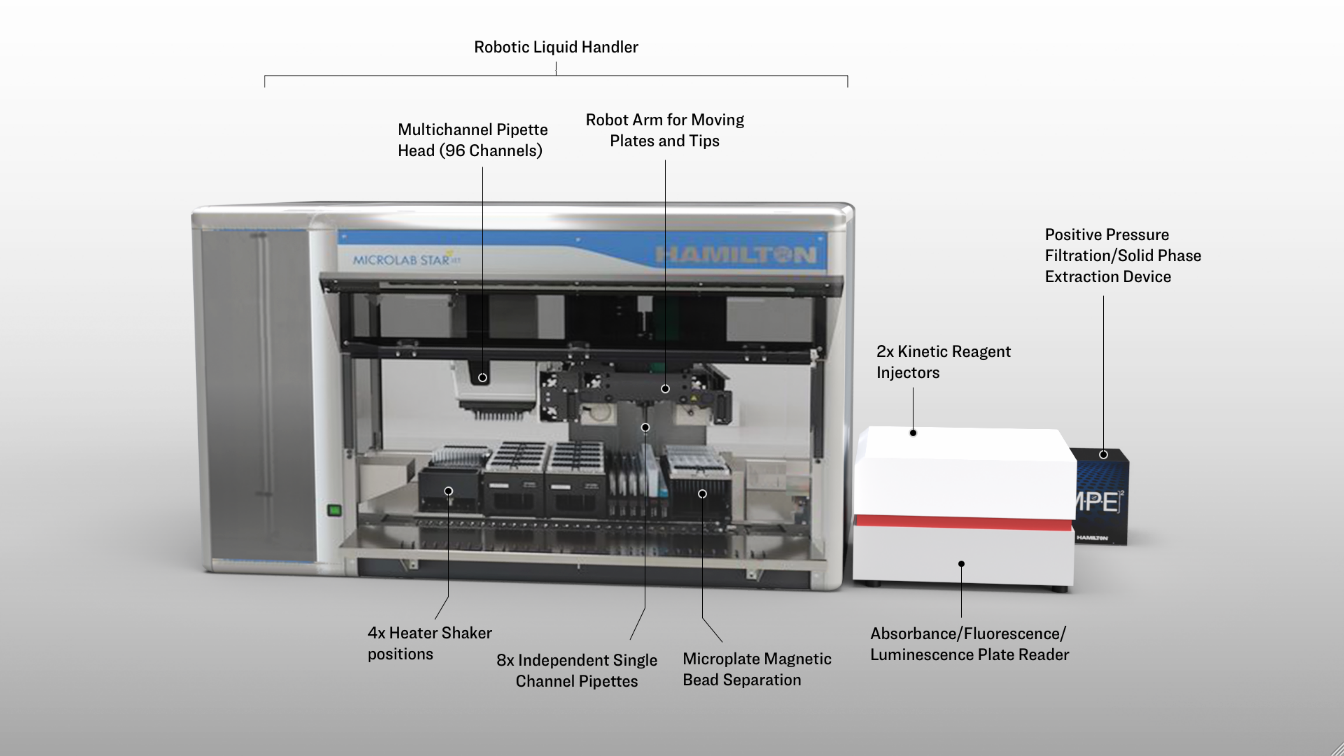
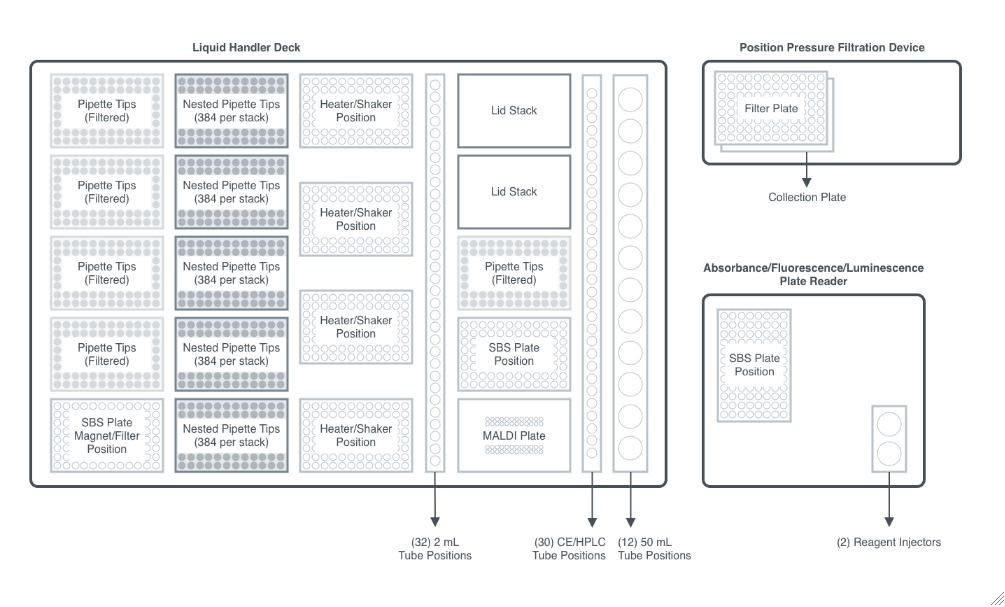
Figure 2.1.1: An overview of the robotic workcell. It is capable of processing plate operations including incubation, shaking, and magnetic bead separation on-deck and filtration and absorbance/fluorescence/luminescence measurement off-deck, with a robot arm facilitating the plate movements.
Super STAR
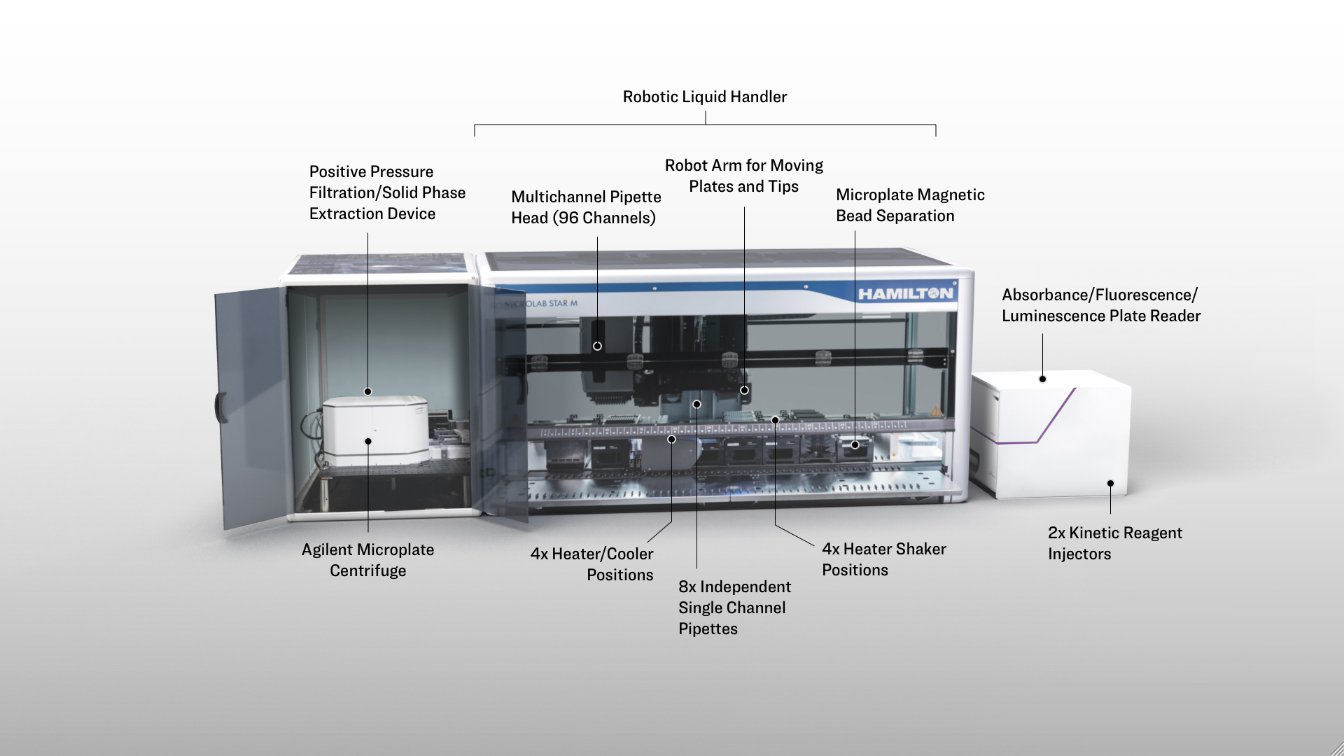
Figure 2.2.1: An overview of the robotic workcell. It is capable of processing plate operations including incubation, shaking, and magnetic bead separation on-deck and centrifugation, filtration, and absorbance/fluorescence/luminescence measurement off-deck, with a robot arm facilitating the plate movements.
Experiment Options
General
ReactionChemistry
Indicates the reactive group that forms the covalent bond between conjugation samples. Null indicates a custom reaction chemistry.
Pattern Description: An object of type or subtype Model[ReactionMechanism] or NHSester, Carbodiimide, or Maleimide or Null.
Programmatic Pattern: ((NHSester | Carbodiimide | Maleimide) | ObjectP[{Model[ReactionMechanism]}]) | Null
ReactantsStoichiometricCoefficients
ProductStoichiometricCoefficient
The number of conjugated molecules created by reaction of input molecules at the ratio indicated by ReactantsStoichiometricCoefficients.
ExpectedYield
The percentage of input molecules that will be converted into conjugated product upon completion of the experiment.
ProcessingOrder
The order for processing the conjugation reactions, where Parallel indicates all samples are processed together from activation to quenching. Parallel processing is ideal for experiments with long reaction times where precision is not a concern and/or when instrumentation that can accommodate all reactions simultaneously is used. Batch indicates certain groups of samples are processed together such that all reactions from activation to quenching are completed for a given group before processing the next group of samples. Grouped processing is ideal for experiments where the instrumentation used cannot accommodate all samples simultaneously. Serial indicates samples are processed sequentially such at all reactions from activation to quenching are completed for a given sample before proceeding to the next sample. Sequential processing is ideal when reaction times are short and must be precise and/or the instruments used can only process one sample at a time.
ProcessingBatchSize
Pattern Description: Greater than or equal to 1 and less than or equal to 1000 in increments of 1 or Null.
ProcessingBatches
The groups of sample pools to process together in the experiment, applicable if ProcessingOrder->Batch.
Pattern Description: List of one or more list of one or more list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample entries entries entries or Null.
Programmatic Pattern: ({{{(ObjectP[{Model[Sample], Object[Sample]}] | _String)..}..}..} | Automatic) | Null
ReactionVessel
The containers in which samples are mixed with ConjugationReactionBuffer and incubated to covalently link the samples. Indices indicate specific grouping of samples if desired. Solid phase sample containers will be prioritized such that solid phase samples are not moved during conjugation reaction assembly.
Default Calculation: Automatically set to the smallest tube that will fit all the reaction components.
Pattern Description: An object of type or subtype Object[Container] or Model[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Object[Container], Model[Container]}] | _String) | {GreaterEqualP[1, 1], ObjectP[{Model[Container], Object[Container]}] | _String}) | Automatic) | Null
PreWash
PreWash
Indicates if the solid phase samples should be washed before the activation and/or conjugation reaction.
Default Calculation: Automatically set to True if any PreWash options are specified, otherwise set to False.
PreWashMethod
Default Calculation: When PreWash is True, automatically set based on PreWash method options. If filtration related options are specified will resolve to Filter, if pellet related options are specified will resolve to Pellet, and if magnetic transfer options are specified will resolve to Magnetic. Otherwise defaults to Pellet.
PreWashNumberOfWashes
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PreWashTime
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 4000 minutes or Null.
PreWashTemperature
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
PreWashBuffer
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
PreWashBufferVolume
Default Calculation: Automatically set to half the sample container volume PreWash is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PreWashMix
PreWashMixType
The method used to mix the samples and wash buffer. Mixing methods Pipette and Invert with occur only at the start of the PreWashTime. All other methods will occur continuously for the duration of the PreWashTime.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
PreWashMixVolume
Default Calculation: Automatically set to half the PreWashBufferVolume if PreWashMixType is Pipette. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PreWashNumberOfMixes
Default Calculation: Automatically set to 5 if PreWashMixType is Pipette or Invert. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PreWashMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
PreWashFiltrationType
Default Calculation: Automatically set to a filtration type appropriate for the volume of sample being filtered when PreWashMethod is Filter.
PreWashInstrument
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
PreWashFilter
Default Calculation: Automatically set to a filter appropriate for the filtration type and instrument if PreWashMethod is Filter.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Object[Container, Plate, Filter], Model[Container, Vessel, Filter], Object[Container, Vessel, Filter], Model[Item, Filter], or Object[Item, Filter] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Plate, Filter], Object[Container, Plate, Filter], Model[Container, Vessel, Filter], Object[Container, Vessel, Filter], Model[Item, Filter], Object[Item, Filter]}] | _String) | Automatic) | Null
PreWashFilterStorageCondition
The conditions under which any filters used for PreWash should be stored after the wash is completed.
PreWashFilterMembraneMaterial
Default Calculation: Will automatically resolve to PES or to the MembraneMaterial of Filter if it is specified.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PreWashPrefilterMembraneMaterial
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PreWashFilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to .22 Micron or to the PoreSize of Filter if it is specified. Will automatically resolve to Null if MolecularWeightCutoff is specified.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PreWashFilterMolecularWeightCutoff
The molecular weight cutoff of the filter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to Null or to the MolecularWeightCutoff of Filter if it is specified.
Pattern Description: 3 kilograms per mole, 3. kilograms per mole, 10 kilograms per mole, 10. kilograms per mole, 30 kilograms per mole, 30. kilograms per mole, 40 kilograms per mole, 40. kilograms per mole, 50 kilograms per mole, 50. kilograms per mole, 100 kilograms per mole, 100. kilograms per mole, 300 kilograms per mole, 300. kilograms per mole, 7 kilograms per mole, 7. kilograms per mole, 40 kilograms per mole, 40. kilograms per mole, or 30.8328 kilograms per mole or Null.
PreWashPrefilterPoreSize
The pore size of the prefilter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to .45 Micron if a prefilter membrane material is specified
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PreWashFiltrationSyringe
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
PreWashSterileFiltration
PreWashFilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane, or a filter plate for vacuum filtration..
Default Calculation: Resolves to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used, or a filter plate for vacuum filtration..
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
PreWashCentrifugationIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if filtering type is Centrifuge or PreWashMethod is Pellet.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
PreWashCentrifugationTime
Default Calculation: Will automatically resolve to 5 Minute if filtering type is Centrifuge or PreWashMethod is Pellet.
PreWashCentrifugationTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during PreWash.
Default Calculation: Will automatically resolve to 22 Celsius if filtering type is Centrifuge or PreWashMethod is Pellet.
PreWashMagnetizationRack
Default Calculation: Automatically set to a magnetized rack that can hold the source/intermediate container, if PreWashMethod->Magnetic is specified.
Pattern Description: An object of type or subtype Model[Container, Rack] or Object[Container, Rack] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Rack], Object[Container, Rack]}] | _String) | Automatic) | Null
PreWashResuspension
Indicates if samples should be resuspended in PreWashResuspensionBuffer after washing and before activation or conjugation reactions.
Default Calculation: Automatically set based on PreWash and prewash resuspension options. If PreWash is False, set to False.
PreWashResuspensionTime
Default Calculation: Automatically, set to 1 Minute when PreWashResuspension is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 4000 minutes or Null.
PreWashResuspensionTemperature
Default Calculation: Automatically, set to Ambient when PreWashResuspension is True. Otherwise, set to Null.
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
PreWashResuspensionDiluent
Default Calculation: Automatically set to 1xPBS if PreWashResuspension is True. Otherwise, set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
PreWashResuspensionDiluentVolume
The volume of the PreWashResuspensionDiluent used to resuspend the sample after the PreWash is complete.
Default Calculation: Automatically set to the original sample volume PreWashResuspension is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PreWashResuspensionMix
Default Calculation: Automatically set to True if PreWashResuspenion is True. Otherwise, set to Null.
PreWashResuspensionMixType
Default Calculation: Automatically, set to Pipette if PreWashResuspensionMix is True. Otherwise, set to Null.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
PreWashResuspensionMixVolume
Default Calculation: Automatically set to half the PreWashResuspensionDiluentVolume if PreWashResuspensionMixType is Pipette. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PreWashResuspensionNumberOfMixes
The number of aspirate and dispense cycles or inversions cycles used to mix the sample during resuspension.
Default Calculation: Automatically set to 5 if PreWashResuspensionMixType is Pipette or Invert. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PreWashResuspensionMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
Activation
Activate
Indicates if functional groups on each of the conjugation samples need to be activated of modified prior to conjugation.
Default Calculation: Automatically set to True if ReactionChemistry is Maleimide and the sample is a protein or if any Activation options are specified.
ActivationReactionVolume
The total volume of the activation reaction including ActivationSampleVolume, ActivationReagentVolume, and ActivationDiluentVolume.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 1000 milliliters or Null.
TargetActivationSampleConcentration
Programmatic Pattern: ((GreaterP[0*(Milligram/Milliliter)] | GreaterP[0*Micromolar]) | Automatic) | Null
ActivationSampleVolume
Default Calculation: When Activate is True, automatically set to the sample volume that results in TargetActivationSampleConcentration when added to ActivationReactionVolume. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 1000 milliliters or Null.
ActivationReagent
The reagent that activates or modifies functional groups on the corresponding sample in preparation for conjugation. For Maleimide reactions, 100X molar excess of TCEP is recommended to reduce protein cysteine disulfide bonds.
Default Calculation: Automatically set to 10mM TCEP in 50mM Tris-HCL pH 7.5 if ReactionChemsitry is Maleimide. TCEP unlike other reducing agents does not contain thiols that may interfere with conjugation, therefore it is not necessary to remove this reagent prior to the maleimide conjugation reaction.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
TargetActivationReagentConcentration
Programmatic Pattern: ((GreaterP[0*(Milligram/Milliliter)] | GreaterP[0*Micromolar]) | Automatic) | Null
ActivationReagentVolume
Default Calculation: When Activate is True, automatically set to the volume of ActivationReagent that results in TargetActivationReagentConcentration when added to ActivationVolume. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 1000 milliliters or Null.
ActivationDiluent
The reagent used to dilute the ActivationSampleVolume + ActivationReagentVolume to the final ActivationReactionVolume.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ActivationDiluentVolume
Default Calculation: When Activate is True, automatically set to the difference in volume between (ActivationVolume + ActivationReagentVolume) and ActivationReactionVolume. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 1000 milliliters or Null.
ActivationContainer
The containers in which each sample is activated by mixing and incubation with ActivationReagent and ActivationDiluent, with indices indicating specific grouping of samples if desired. If Null, the activation reaction will occur in the sample's current container.
Pattern Description: An object of type or subtype Object[Container] or Model[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: ((ObjectP[{Object[Container], Model[Container]}] | _String) | {GreaterEqualP[1, 1], ObjectP[{Model[Container], Object[Container]}] | _String}) | Null
ActivationTime
The length of the activation reaction incubation. To mix the activation reagent and sample immediately prior to addition to the conjugation reaction set this value to zero.
Default Calculation: Automatically, set to 15 Minutes when Activate is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 4000 minutes or Null.
ActivationTemperature
Default Calculation: Automatically, set to Ambient when Activate is True and ActivationTime is greater than zero. Otherwise, set to Null.
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
ActivationMix
ActivationMixType
The method used to mix the activation reaction before or during incubation. Mixing methods Pipette and Invert with occur only at the start of the ActivationTime. All other methods will occur continuously for the duration of the ActivationTime.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
ActivationMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
ActivationMixVolume
Default Calculation: Automatically, set to half the activation reaction volume if ActivationMixType is Pipette. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 50000 microliters or Null.
ActivationNumberOfMixes
Default Calculation: Automatically, set to 5 if ActivationMixType is Pipette or Invert. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PostActivationWash
Indicates if samples should be washed after the activation reaction and before the conjugation reaction.
Default Calculation: Automatically set to True if Activate is True and the sample is a solid phase support. Otherwise, set to Null.
PostActivationWashMethod
Default Calculation: If PostActivationWash is True, automatically set based on method specific PostActivationWash options.
PostActivationNumberOfWashes
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PostActivationWashTime
Default Calculation: Automatically, set to 1 Minute when PostActivationWash is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 4000 minutes or Null.
PostActivationWashTemperature
Default Calculation: Automatically, set to Ambient when PostActivationWash is True. Otherwise, set to Null.
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
PostActivationWashBuffer
Default Calculation: Automatically set to 1X PBS if PostActivationWash is True. Otherwise, set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
PostActivationWashBufferVolume
The volume of the PostActivationWashBuffer used to wash the sample after the activation reaction is complete.
Default Calculation: Automatically set to 200 Microliter PostActivationWash is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostActivationWashMix
Default Calculation: Automatically set to True if PostActivationWash is True. Otherwise, set to Null.
PostActivationWashMixType
The method used to mix the samples and wash buffer. Mixing methods Pipette and Invert with occur only at the start of the PostActivationWashTime. All other methods will occur continuously for the duration of the PostActivationWashTime.
Default Calculation: Automatically, set to Pipette if PostActivationWash is True. Otherwise, set to Null.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
PostActivationWashMixVolume
Default Calculation: Automatically set to half the PostActivationWashBufferVolume if PostActivationWashMixType is Pipette. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostActivationWashNumberOfMixes
The number of aspirate and dispense cycles or inversion cycles used to mix the sample during washing.
Default Calculation: Automatically set to 5 if PostActivationWashMixType is Pipette or Invert. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PostActivationWashMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
PostActivationWashFiltrationType
Default Calculation: Automatically set to a filtration type appropriate for the volume of sample being filtered when PostActivationWashMethod is Filter.
PostActivationWashInstrument
Default Calculation: Will automatically resolve to an instrument appropriate for the PostActivationWashMethod.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
PostActivationWashFilter
Default Calculation: Automatically set to a filter appropriate for the filtration type and instrument if PostActivationWashType is Filter.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Object[Container, Plate, Filter], Model[Container, Vessel, Filter], Object[Container, Vessel, Filter], Model[Item, Filter], or Object[Item, Filter] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Plate, Filter], Object[Container, Plate, Filter], Model[Container, Vessel, Filter], Object[Container, Vessel, Filter], Model[Item, Filter], Object[Item, Filter]}] | _String) | Automatic) | Null
PostActivationWashFilterStorageCondition
The conditions under which any filters used for PostActivationWash should be stored after the wash is completed.
PostActivationWashFilterMembraneMaterial
Default Calculation: Will automatically resolve to PES or to the MembraneMaterial of Filter if it is specified.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PostActivationWashPrefilterMembraneMaterial
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PostActivationWashFilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to .22 Micron or to the PoreSize of Filter if it is specified. Will automatically resolve to Null if MolecularWeightCutoff is specified.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PostActivationWashFilterMolecularWeightCutoff
The molecular weight cutoff of the filter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to Null or to the MolecularWeightCutoff of Filter if it is specified.
Pattern Description: 3 kilograms per mole, 3. kilograms per mole, 10 kilograms per mole, 10. kilograms per mole, 30 kilograms per mole, 30. kilograms per mole, 40 kilograms per mole, 40. kilograms per mole, 50 kilograms per mole, 50. kilograms per mole, 100 kilograms per mole, 100. kilograms per mole, 300 kilograms per mole, 300. kilograms per mole, 7 kilograms per mole, 7. kilograms per mole, 40 kilograms per mole, 40. kilograms per mole, or 30.8328 kilograms per mole or Null.
PostActivationWashPrefilterPoreSize
The pore size of the Prefilter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to .45 Micron if a Prefilter membrane material is specified
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PostActivationWashFiltrationSyringe
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
PostActivationWashSterileFiltration
PostActivationWashFilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane, or a filter plate for vacuum filtration..
Default Calculation: Resolves to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used, or a filter plate for vacuum filtration..
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
PostActivationWashCentrifugationIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if filtering type is Centrifuge or PostActivationWashMethod is Pellet.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
PostActivationWashCentrifugationTime
Default Calculation: Will automatically resolve to 5 Minute if filtering type is Centrifuge or PostActivationWashMethod is Pellet.
PostActivationWashCentrifugationTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during PostActivationWash.
Default Calculation: Will automatically resolve to 22 Celsius if filtering type is Centrifuge or PostActivationWashMethod is Pellet.
PostActivationWashMagnetizationRack
The magnetized rack that the source/intermediate container will be placed in during the PostActivationWash.
Default Calculation: Automatically set to a magnetized rack that can hold the source/intermediate container, if PostActivationWashMethod->Magnetic is specified.
Pattern Description: An object of type or subtype Model[Container, Rack] or Object[Container, Rack] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Rack], Object[Container, Rack]}] | _String) | Automatic) | Null
PostActivationResuspension
Indicates if samples should be resuspended in PostActivationResuspensionDiluent after washing and before the conjugation reaction.
PostActivationResuspensionTime
Default Calculation: Automatically, set to 1 Minute when PostActivationResuspension is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 4000 minutes or Null.
PostActivationResuspensionTemperature
Default Calculation: Automatically, set to Ambient when PostActivationResuspension is True. Otherwise, set to Null.
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
PostActivationResuspensionDiluent
Default Calculation: Automatically set to 1X PBS if PostActivationWash is True. Otherwise, set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
PostActivationResuspensionDiluentVolume
The volume of the PostActivationResuspensionDiluent used to resuspend the sample after the PostActivationWash is complete.
Default Calculation: Automatically set to 200 Microliter PostActivationWash is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostActivationResuspensionMix
Default Calculation: Automatically set to True if PostActivationWash is True. Otherwise, set to Null.
PostActivationResuspensionMixType
Default Calculation: Automatically, set to Pipette if PostActivationResuspensionMix is True. Otherwise, set to Null.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
PostActivationResuspensionMixVolume
The volume used to mix the samples by pipetting after dispensing of the PostActivationResuspensionDiluent.
Default Calculation: Automatically set to half the PostActivationResuspensionDiluentVolume if PostActivationResuspensionMixType is Pipette. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostActivationResuspensionNumberOfMixes
The number of aspirate and dispense cycles or inversion cycles used to mix the sample during resuspension.
Default Calculation: Automatically set to 5 if PostActivationResuspensionMixType is Pipette or Invert. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PostActivationResuspensionMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
ActivationSamplesOutStorageCondition
The conditions under which any leftover samples from the activation reaction are stored after the samples are transferred to the ConjugationContainer.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Conjugation
ConjugationReactionVolume
The total volume of the conjugation reaction including ConjugationAmount and ConjugationReactionBufferVolume.
Default Calculation: Automatically, set to the full volume of the conjugate samples or 500 uL if all samples are solid.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters.
TargetConjugationSampleConcentration
Default Calculation: Automatically set to the concentration given by ConjugationAmount/ConjugationReactionVolume.
Programmatic Pattern: ((GreaterP[0*(Milligram/Milliliter)] | GreaterP[0*Micromolar]) | Automatic) | Null
ConjugationAmount
The volume or mass of each sample that is used in the conjugation reaction. By default the entire volume or mass of the sample is used. If ConjugationAmount is All and the sample is solid, the aqueous samples and reagents for the conjugation reaction will be transferred into the solid sample container.
Pattern Description: All or greater than or equal to 0 micrograms and less than or equal to 50 grams or greater than or equal to 0 microliters and less than or equal to 50 milliliters.
ConjugationReactionBuffer
Indicates the reagent used to dilute or resuspend the conjugate sample during the conjugation reaction.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ConjugationReactionBufferVolume
The volume of the ConjugationReactionBuffer that is mixed with the corresponding conjugation samples to assemble the conjugation reaction.
Default Calculation: Automatically, set to the conjugation reaction volume such that the sample is diluted 2-fold.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
ConcentratedConjugationReactionBuffer
Indicates the concentrated reagent used to dilute or resuspend the conjugate sample during the conjugation reaction.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ConcentratedConjugationReactionBufferDilutionFactor
The relative amount that the ConcentratedConjugateBuffer should be diluted prior to use in the conjugation reaction.
ConjugationReactionBufferDiluent
The diluent used to decrease the ConcentratedConjugationBuffer concentration by ConjugationBufferDilutionFactor prior to use in the conjugation reaction.
Default Calculation: If ConcentratedConjugationReactionBuffer is specified, automatically resolves to 1x PBS.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ConjugationDiluentVolume
The volume of the ConjugationBufferDiluent that is mixed with ConcentratedConjugateBuffer prior to use in the conjugation reaction.
Default Calculation: Automatically set to the ConjugationBufferVolume*(1 - ConjugationReactionBufferVolume/ConcentratedConjugationReactionBufferDilutionFactor).
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
ConjugationTime
Default Calculation: Automatically set based on the reaction chemistry to a literature-based recommended value.
ConjugationTemperature
Default Calculation: Automatically set based on the reaction chemistry to a literature-based recommended value.
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius.
ConjugationMix
ConjugationMixType
The method used to mix the samples for conjugation. Mixing methods Pipette and Invert with occur only at the start of the ConjugationTime. All other methods will occur continuously for the duration of the ConjugationTime.
Default Calculation: Automatically, set to based on mix options when ConjugationMix is True. Otherwise, set to Null.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
ConjugationMixVolume
Default Calculation: Automatically set to half the ConjugationReactionVolume if ConjugationMixType is Pipette. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
ConjugationNumberOfMixes
The number of aspirate and dispense or inversion cycles used to mix the sample at the start of the conjugation reaction.
Default Calculation: Automatically set to 5 if ConjugationMixType is Pipette or Invert. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
ConjugationMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
Quenching
QuenchConjugation
PredrainReactionMixture
Indicates if the aqueous reaction reagents should be removed from any solid phase samples or precipitates after the conjugation reaction is complete to prepare the sample for quenching or subsequent workup.
PredrainMethod
The method used to separate the aqueous and solid reaction components when PredrainReactionMixture is True.
Default Calculation: Automatically set to Pellet if PredrainReactionMixture is True. Otherwise set to Null.
PredrainInstrument
Default Calculation: Will automatically resolve to an instrument appropriate for the PredrainMethod.
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
PredrainCentrifugationIntensity
The rotational speed or force at which the samples will be centrifuged during PredrainReactionReagents.
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if PredrainMethod is Pellet.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
PredrainCentrifugationTime
PredrainTemperature
The temperature at which the samples will be held during centrifugation or magnetic separation when PredrainReactionReagents is True.
PredrainMagnetizationRack
The magnetized rack that the source/intermediate container will be placed in during the PredrainReactionMixture.
Default Calculation: Automatically set to a magnetized rack that can hold the source/intermediate container, if PredrainMethod->Magnetic is specified.
Pattern Description: An object of type or subtype Model[Container, Rack] or Object[Container, Rack] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Rack], Object[Container, Rack]}] | _String) | Automatic) | Null
PredrainMagnetizationTime
The time that the source sample should be left on the magnetic rack until the magnetic components are settled at the side of the container.
Default Calculation: Automatically set to 15 Second if PredrainMethod->Magnetic. Otherwise, set to Null.
QuenchReactionVolume
The total volume of the quenching reaction including the ConjugationReactionVolume + QuenchReagentVolume.
Default Calculation: Automatically, based on the QuenchReagentDilutionFactor. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 1000 milliliters or Null.
QuenchReagent
The reagent that should be used to stop the conjugation reaction and render any unbound functional groups inactive.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
QuenchReagentDilutionFactor
The relative amount that the QuenchReagent should be diluted in QuenchReaction to reach its final active concentration.
QuenchReagentVolume
Default Calculation: Automatically, set to 1 uL QuenchReagent when QuenchConjugation is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 1000 milliliters or Null.
QuenchTime
Default Calculation: Automatically, set to 5 Minutes when QuenchConjugation is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 4000 minutes or Null.
QuenchTemperature
Default Calculation: Automatically, set to Ambient QuenchConjugation is True. Otherwise, set to Null.
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
QuenchMix
QuenchMixType
The method used to mix the samples with QuenchReagent. Mixing methods Pipette and Invert with occur only at the start of the QuenchTime. All other methods will occur continuously for the duration of the QuenchTime.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
QuenchMixVolume
The volume used to mix the sample and QuenchReagent by pipetting at the start of the quench reaction.
Default Calculation: Automatically set to half the QuenchReactionVolume if QuenchMixType is Pipette. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
QuenchNumberOfMixes
The number of aspirate and dispense or inversion cycles used to mix the sample at the start of the quench reaction.
Default Calculation: Automatically set to 5 if QuenchMixType is Pipette or Invert. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
QuenchMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
Post-Conjugation Workup
PostConjugationWorkup
PostConjugationWorkupMethod
PostConjugationWorkupBuffer
Indicates the reagent used to process the sample after the conjugation and quenching reactions are complete.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
PostConjugationWorkupBufferVolume
The volume of the PostConjugationWashBuffer used to wash the sample after the conjugation reaction is complete.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostConjugationWorkupIncubationTime
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 4000 minutes or Null.
PostConjugationWorkupIncubationTemperature
Pattern Description: Ambient or greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
PostConjugationWorkupMix
Default Calculation: Automatically set to True if PostConjugationWorkupBuffer is specified. Otherwise, set to False.
PostConjugationWorkupMixType
Default Calculation: Automatically, set to Pipette if PostConjugationWorkup is True. Otherwise, set to Null.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
PostConjugationWorkupMixVolume
Default Calculation: Automatically set to half the sample volume if PostConjugationWorkupMix is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostConjugationWorkupNumberOfMixes
The number of aspirate and dispense cycles or inversion cycles used to mix the sample prior to PostConjugationWorkupIncubationTime.
Default Calculation: Automatically set to 5 if PostConjugationWorkupMix is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PostConjugationWorkupMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
PostConjugationFiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
PostConjugationWorkupInstrument
Default Calculation: Will automatically resolve to an instrument appropriate for the PostConjugationWorkupMethod.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
PostConjugationFilter
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Object[Container, Plate, Filter], Model[Container, Vessel, Filter], Object[Container, Vessel, Filter], Model[Item, Filter], or Object[Item, Filter] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Plate, Filter], Object[Container, Plate, Filter], Model[Container, Vessel, Filter], Object[Container, Vessel, Filter], Model[Item, Filter], Object[Item, Filter]}] | _String) | Automatic) | Null
PostConjugationFilterStorageCondition
The conditions under which any filters used by this experiment should be stored after the protocol is completed.
PostConjugationFilterMembraneMaterial
Default Calculation: Will automatically resolve to PES or to the MembraneMaterial of Filter if it is specified.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PostConjugationPrefilterMembraneMaterial
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PostConjugationFilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to .22 Micron or to the PoreSize of Filter if it is specified. Will automatically resolve to Null if MolecularWeightCutoff is specified.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PostConjugationFilterMolecularWeightCutoff
The molecular weight cutoff of the filter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to Null or to the MolecularWeightCutoff of Filter if it is specified.
Pattern Description: 3 kilograms per mole, 3. kilograms per mole, 10 kilograms per mole, 10. kilograms per mole, 30 kilograms per mole, 30. kilograms per mole, 40 kilograms per mole, 40. kilograms per mole, 50 kilograms per mole, 50. kilograms per mole, 100 kilograms per mole, 100. kilograms per mole, 300 kilograms per mole, 300. kilograms per mole, 7 kilograms per mole, 7. kilograms per mole, 40 kilograms per mole, 40. kilograms per mole, or 30.8328 kilograms per mole or Null.
PostConjugationPrefilterPoreSize
The pore size of the prefilter; all particles larger than this should be removed during the filtration.
Default Calculation: Will automatically resolve to .45 Micron if a prefilter membrane material is specified
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PostConjugationFiltrationSyringe
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
PostConjugationSterileFiltration
PostConjugationFilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane, or a filter plate for vacuum filtration.
Default Calculation: Resolves to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used, or a filter plate for vacuum filtration.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
PostConjugationCentrifugationIntensity
The rotational speed or force at which the samples will be centrifuged during PostConjugationWorkup.
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if filtering type is Centrifuge or PostConjugationWorkupMethod is Pellet.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
PostConjugationCentrifugationTime
Default Calculation: Will automatically resolve to 5 Minute if filtering type is Centrifuge or PostConjugationWorkupMethod is Pellet.
PostConjugationCentrifugationTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during PostConjugationWorkup.
Default Calculation: Will automatically resolve to 22 Celsius if filtering type is Centrifuge or PostConjugationWorkupMethod is Pellet.
PostConjugationMagnetizationRack
The magnetized rack that the source/intermediate container will be placed in during the PostConjugationWorkup.
Default Calculation: Automatically set to a magnetized rack that can hold the source/intermediate container, if PostConjugationWorkupMethod->Magnetic is specified.
Pattern Description: An object of type or subtype Model[Container, Rack] or Object[Container, Rack] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Rack], Object[Container, Rack]}] | _String) | Automatic) | Null
PostConjugationResuspension
Indicates whether the sample should be resuspended in PostConjugationResuspensionDiluent after PostConjugationWorkup is complete.
Default Calculation: Automatically set to True if any resuspension options are specified. Otherwise set to False.
PostConjugationResuspensionDiluent
Default Calculation: Automatically set to 1X PBS if PostConjugationResuspension is True. Otherwise, set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
PostConjugationResuspensionDiluentVolume
The volume of the PostConjugationResuspensionDiluent used to resuspend the sample after the PostConjugationWash is complete.
Default Calculation: Automatically set to 200 Microliter PostConjugationResuspension is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostConjugationResuspensionMix
Default Calculation: Automatically set to True if PostConjugationResuspension is True. Otherwise, set to Null.
PostConjugationResuspensionMixType
Default Calculation: Automatically, set to Pipette if PostConjugationResuspensionMix is True. Otherwise, set to Null.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
PostConjugationResuspensionMixRate
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 10000 revolutions per minute or Null.
PostConjugationResuspensionMixVolume
Default Calculation: Automatically set to half the PostConjugationResuspensionDiluentVolume if PostConjugationResuspensionMix is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 1000000 microliters or Null.
PostConjugationResuspensionNumberOfMixes
Default Calculation: Automatically set to 5 if PostConjugationResuspensionMix is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PostConjugationResuspensionMixUntilDissolved
Indicates if the sample should be mixed in an attempt to completed dissolve any solid components following addition of liquid.
Default Calculation: Automatically set to True if MaxMixTime or MaxNumberOfMixes are specified, or False otherwise.
PostConjugationResuspensionMaxNumberOfMixes
Default Calculation: Automatically set to 50 if PostConjugationResuspensionMixUntilDissolved is True. Otherwise, set to Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 100 in increments of 1 or Null.
PostConjugationResuspensionTime
Default Calculation: Automatically set to 30 minutes unless MixType is set to Pipette or Invert, in which case it is set to Null.
PostConjugationResuspensionMaxTime
The maximum duration for which the samples should be mixed/incubated in an attempt to dissolve any solid components following addition of liquid.
Default Calculation: Automatically set based on the MixType if PostConjugationResuspensionMixUntilDissolved is set to True. If PostConjugationResuspensionMixUntilDissolved is False, resolves to Null.
PostConjugationResuspensionTemperature
Pattern Description: Ambient or greater than or equal to 22 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[22*Celsius, $MaxIncubationTemperature] | Ambient) | Automatic) | Null
Post Experiment
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
PreparatoryPrimitives
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more a primitive with head Define, Transfer, Mix, Aliquot, Consolidation, FillToVolume, Incubate, Filter, Wait, Centrifuge, or Resuspend entries or Null.
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentratedBufferDilutionFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or list of one or more any well from A1 to H12 or any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..} | {(((Automatic | Null) | WellPositionP) | {((Automatic | Null) | WellPositionP)..})..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries or list of one or more list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries entries or list of one or more list of one or more Automatic or Null or {Index, Container} entries entries.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..} | {{((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..}..} | {{({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
ImageSample
Indicates if any samples that are modified in the course of the experiment should be freshly imaged after running the experiment. Please note that public samples are imaged regardless of the value of this option.
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Example Calls
Conjugate molecules in the desired samples.
Conjugation of analytes contained in two samples where the resulting molecule has the properties contained in myConjugatedMoleculeIdentityModel.

Conjugate molecules in the desired samples overnight at room temperature.
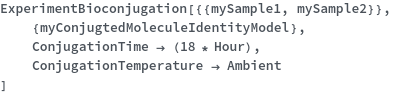
Perform the following conjugation steps: sample activation, conjugation, and conjugation reaction quenching. First, the sample is modified to activate functional groups in preparation for conjugation with the paired sample. Next, the samples are mixed and incubated to create a checmically linked product. Lastly, a quencher is added to terminate the conjugation reaction.

Perform sample workup after conjugation is complete.

Warnings and Errors
Messages (40)
ConflictingPostActivationResuspensionMixOptions (1)
ConflictingPostActivationResuspensionOptions (1)
ConflictingPostActivationWashMixOptions (1)
ConflictingPostConjugationMixUntilDissolvedOptions (1)
ConflictingPostConjugationResuspensionMixOptions (1)
ConflictingPostConjugationResuspensionOptions (1)
ConflictingPostConjugationWorkupMixOptions (1)
ConflictingPostConjugationWorkupOptions (1)
ConflictingPreWashResuspensionMixOptions (1)
InvalidActivationReagentVolumes (1)
InvalidConjugationMixVolumes (1)
InvalidPostConjugationOptions (1)
InvalidPostConjugationResuspensionDiluentVolumes (1)
InvalidPostConjugationResuspensionMixVolumes (1)
InvalidQuenchingOptions (1)
InvalidQuenchMixVolumes (1)
InvalidReactionVessel (1)
ObjectDoesNotExist (2)
TooManyConjugationBufferOptions (1)
TooManyConjugationBuffers (1)
UnknownProductStoichiometry (1)
Possible Issues
Last modified on Thu 11 Apr 2024 17:15:07