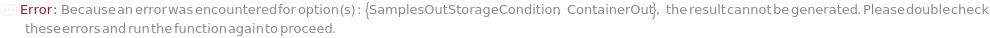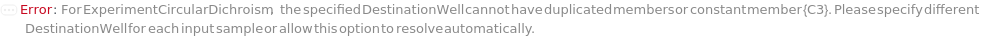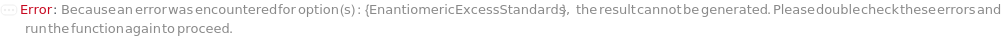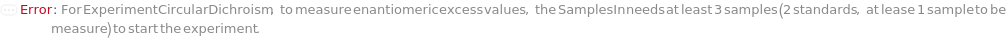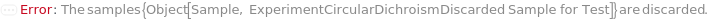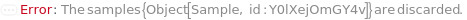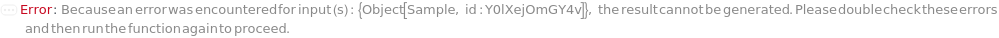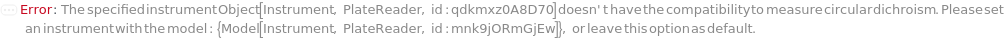ExperimentCircularDichroism
ExperimentCircularDichroism[Samples]⟹Protocol
generates a Protocol object for measuring the differential absorption of left and right circularly polarized light of the Samples.
Circular Dichroism (CD, aka Electronic Circular Dichroism (ECD)) measures how samples differentially absorb left and right circularly polarized light radiation, either at a specific wavelength or over a range of wavelengths. In UV-Vis experiment (e.g., ExperimentAbsorbanceSpectroscopy, ExperimentUVMelting etc), samples are exposed to the unpolarized light, where its electric and magnetic fields perpendicular oscillate in all directions perpendicular to the propagation direction of the light. In circularly polarized light, the electric field vector rotates around the propagation direction at a constant magnitude, resulting in the electric field vector appearing to trace a circle throughout one wave frequency. Optically active chiral molecules will preferentially absorb one direction of the circularly polarized light. The difference in absorption of the left and right circularly polarized light can be measured and quantified as absorption bands. CD spectroscopy can probe the secondary structure composition of polypeptides/proteins and determine the tertiary structure class of globular proteins.
Experimental Principles
Figure 1.1: Procedural overview of a CircularDichroism experiment using an Ekko CD plate reader.Step 1:Samples and blanks are loaded to the plate for reading.Step 2 (optional):the sample volume will be measured by the supersonic method to calculate the pathlength.Step 3:Samples are loaded to the instrument.Step 4:The circular dichroism of samples is measured as ellipticity,based on differential absorption of the left and right circularly polarized light of samples.Step 5:Data is plotted and visualized.
Instrumentation
Ekko
Figure 2.1.1: In an Ekko CD Microplate Reader, a Xenon Lamp emits the full-spectrum light. The light is then split and diffracted into several beams by passing a grating monochromator to emit non-polarized light. The non-polarized light then passes the polarizer to generate linear polarized light. The linear polarized light then passes through the photoelastic modulator to create both left- and right-hand circular polarized light. Finally, the differential absorbed circularly polarized light. The transmitted light is then directed through a prism that splits the beam into wavelengths from 185 nm to 900 nm as measured by the detector.
Experiment Options
General
Instrument
The plate reader that is used for measuring circular dichroism spectroscopy or circular dichroism intensity at specific wavelengths.
Pattern Description: An object of type or subtype Model[Instrument, PlateReader] or Object[Instrument, PlateReader]
PreparedPlate
Indicates if the plate containing the samples for the CircularDichroism experiment has been previously prepared that does not need to run sample preparation steps.
Optics
RetainCover
Indicates if the plate seal or lid on the assay container is left on during measurement to decrease evaporation. It is strongly recommended not to retain a cover because Circular Dichroism can only read from top.
DetectionWavelength
Default Calculation: Automatically resolves based on the EnantiomericExcessWavelength first: If EnantiomericExcessWavelength is specified, resolved to be the same as EnantiomericExcessWavelength. If EnantiomericExcessWavelength is blank, this option resolves based on Analytes option: if Analytes are specified as biomacromolecules (Proteins, Peptides, Oligomers), resolves this option to Span[200 Nanometer, 400 Nanometer]; Else resolved to Span[300 Nanometer, 800 Nanometer].
Programmatic Pattern: (RangeP[185*Nanometer, 850*Nanometer] | RangeP[185*Nanometer, 850*Nanometer] ;; RangeP[185*Nanometer, 850*Nanometer] | {RangeP[185*Nanometer, 850*Nanometer]..}) | Automatic
StepSize
This option determines how often the spectrophotometer will record an circular dichroism absorbance measurement, if the sample was scanned in a range of wavelength. For example, a step size of 2 nanometer indicates that the spectrophotometer will collect circular dichroism absorbance data for every 2 nanometer within the wavelength range.
Default Calculation: Automatically resolves to 1 Nanomer if the DetectionWavelength is resolved as a range values. Otherwise will resove to Null.
Pattern Description: Greater than or equal to 0.5 nanometers and less than or equal to 5 nanometers or Null.
Sample Preparation
ReadPlate
The plate that is loaded with input samples then inserted into the CircularDichroism plater reader instrument.
Default Calculation: Is automatically set to Model[Container, Plate, "Hellma Black Quartz Microplate"].
Pattern Description: An object of type or subtype Model[Container, Plate] or Object[Container, Plate] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Container, Plate], Object[Container, Plate]}] | _String) | Automatic
NumberOfReplicates
The number of times to repeat absorbance reading on each provided sample. If Aliquot -> True, this also indicates the number of times each provided sample will be aliquoted.
MoatSize
Indicates the number of concentric perimeters of wells to leave as empty or filled with MoatBuffer. Quartz plates have high birefringence in the outer ring of wells. So the circular dichroism data collected in the outer ring of wells have noise levels. With MoatSize == 2 (default value), the protocol will ignore the outer ring of wells in the plate.
Figure 3.1: Use the moat options, MoatBuffer, MoatVolume and MoatSize to create an outer ring of wells (moat wells) filled with buffer or leave as empty wells. For quartz plate, since the plate's outer ring is not recommended to be sued for circular dichroism. Empty moat well will auto prevent using those wells. Moat wells with the buffer have been shown to decrease evaporation during long reads.
Default Calculation: Automatically set to 2 if ReadPlate is made by quartz (e.g. Model[Container, Plate, "Hellma Black Quartz Microplate"] and Object[Container,Plate] with this Model). Otherwise will resolve to 0.
MoatBuffer
Indicates the sample to use to fill each moat well. If the moat well is intended to be empty, specify this option as Null.
Default Calculation: Automatically set to Null if RetainCover is False. Otherwise resolve to automatically Milli-Q water.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
MoatVolume
Default Calculation: Automatically set to the minimum volume of the assay plate if MoatBuffer is specified. Otherwise resolves to 0.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 300 microliters or Null.
SampleVolume
Default Calculation: Is automatically set based on the smallest volume value of all SamplesIn and the RecommendedFillVolume of the ReadPlate, whichever is smaller.
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 2000 microliters.
CalculatingMolarEllipticitySpectrum
Decide if the data will be transfered to molar ellipticity after the protocol is finished. If True, the Analyte and AnalyteConcentration option will set a single value for each of SamplesIn.
Default Calculation: If the analyte concentration is specified, this option resolved to True, otherwise resolve to False.
Data Processing
NitrogenPurge
Indicates if the experiment is run under purge with dry nitrogen gas to avoid condensation of ozone generated by the light source.
EmptyAbsorbance
Indicates if a empty well will be scan to check the container's backgound signal at the begining of the experiment.
BlankAbsorbance
Indicates if blank samples are prepared to account for the background signal when reading absorbance of the assay samples.
Blanks
The source used to generate a blank sample whose absorbance is subtracted as background from the absorbance readings of the input sample.
Default Calculation: Automatically set to Null if BlankAbsorbance is False, otherwise set to Model[Sample, "Milli-Q water"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BlankVolumes
The volume of the blank that should be transferred out and used for blank measurements. Set BlankVolumes to Null to indicate blanks should be read inside their current containers.
Default Calculation: Automatically set to Null if BlankAbsorbance is False or to the SampleVolume of the samples if BlankAbsorbance is True.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 300 microliters or Null.
ReadDirection
Indicate the plate path the instrument will follow as it measures circular dichroism absorbance in each well, for instance reading all wells in a row before continuing on to the next row (Row).
Figure 3.2: Use the ReadDirection option to control how the plate reader scans the plate during each read cycle. Choosing a more efficient path will lead to a shorter DetectionInterval. Also note that you may want to consider dripping injectors if you're working with low viscosity injection samples.
AverageTime
The time on data collection for each measurment points. The collected data are averaged for this period of time and export as the result data point for this wavelength.
Pattern Description: Greater than or equal to 0.06 seconds and less than or equal to 18 seconds or Null.
Enatiomeric Excess Measurement
EnantiomericExcessMeasurement
Default Calculation: Automatically set to False. Unless EnantiomericExcessWavelength or EnantiomericExcessStandards is specified by users.
EnantiomericExcessWavelength
Default Calculation: Automatically set to Null if EnantiomericExcessMeasurement is set to False. For EnantiomericExcessMeasurement is True, If DetectionWavelength is specified as a list of single wavelengths, automatically set to the samme as DetectionWavelengh. Else, automatically set to the shortest wavelength specified in the input samples' ExtinctionCoefficients field, and 260 Nanometer if that field is not populated.
Programmatic Pattern: ((RangeP[185*Nanometer, 850*Nanometer] | {RangeP[185*Nanometer, 850*Nanometer]..}) | Automatic) | Null
EnantiomericExcessStandards
Indicate samples with a known EnantiomericExcess values and the corresponding value. Preferrable to have optical pure isomers (with +100% and -100% in enantiomeric excess) and one racemic sample (0% in enantiomeric excess). The blank sample can be used as the racemic sample. Will throw an error message if not enough sample (<=3) is specified as SamplesIn.
Default Calculation: Automatically set to Null if EnantiomericExcessMeasurement is set to False. If EnantiomericExcessMeasurement is True, the first two SamplesIn will be set as Sample, with -100% and 100% Enantiomeric Excess Value, respectively. The blank sample (if any) or the 3rd SamplesIn will be set as the Sample with 0% Enantiomeric Excess Value.
Programmatic Pattern: ({{(ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic, RangeP[-100*Percent, 100*Percent] | Automatic}..} | Automatic) | Null
Sample Information
Analyte
The compound of interest that is present in the given samples, will be used to determine the other settings for the plate reader (e.g. DetectionWavelength) and will be used to caluclate the molar epplicity.
Default Calculation: If populated, will set to the fist user-specified Analyte field in the Object[Sample]. Otherwise, will set to the fist larger compounds in the sample, in the order of Proteins, Peptides, Oligomers, then other small molecules. Otherwise, set Null.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AnalyteConcentrations
Post Experiment
ContainerOut
Default Calculation: Is automatically set to Null if the SamplesOutStorageCondition is set to Disposal. If the SamplesOutStorageCondition is not Disposal, this option resolves based on the reusability of the ReadPlate: if the ReadPlate is reusable (Reusable->True for its Model), this option resolved to be Null, otherwise is resolved to Model[Container,Plate,"96-well 1mL Deep Well Plate"].
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic) | Null
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Model Input
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Programmatic Pattern: ((Null | (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All)) | Automatic) | Null
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise, automatically set to Model[Container, Plate, "Hellma Black Quartz Microplate"].
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSamplePreparation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentratedBufferDilutionFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Basics
Use BlankAbsorbance and EmptyAbsorbance options to generate protocol without blank and and empty well scans.
Molar Ellipticity
The resulting spectra are in ellipticity (mdeg). To generate data in molar ellipticity, specify the CalculatingMolarEllipticitySpectrum, Analyte, AnalyteConcentration in order to auto calculate the molar ellipticity in resulting spectra.
Enantiomeric Excess
To calculate the enantiomeric excess of an unknown sample, submit two standard samples and the unknown samples. Specify each standard sample with their known enantiomeric excess value in the EnantiomericExcessStandards option and specify which wavelengths should be used to calculate the enantiomeric excess in unknown samples EnantiomericExcessWavelengths option.
Preferred Input Containers
Warnings and Errors
Messages (27)
CircularDichroismContainerOutStorageConditionMismatch (1)
CircularDichroismDetectionWavelengthIncompatibleForReadPlate (1)
CircularDichroismEnantiomericExcessWavelengthsNotCovered (1)
CircularDichroismInconsistentAnalyteConcentration (1)
CircularDichroismInvalidDestinationWellLength (1)
CircularDichroismInvalidDetectionWavelength (1)
CircularDichroismNeedMoreEnatiomericExcessStandards (1)
CircularDichroismNeedMoreSamplesForEnantiomericExcessMeasurement (1)
CircularDichroismQuartzPlateCannotRetainCover (1)
CircularDichroismRequiredBlankOption (1)
CircularDichroismTooManySamples (1)
CircularDichroismUnknownEnatiomericExcessStandards (1)
CircularDichroismUnNeededBlankOption (1)
CircularDirchroismQuartzPlateNeedMoat (1)
NeedConcentrationForMolarEllipticity (1)
ObjectDoesNotExist (6)
Do NOT throw a message if we have a simulated container but a simulation is specified that indicates that it is simulated:
Do NOT throw a message if we have a simulated sample but a simulation is specified that indicates that it is simulated:
Throw a message if we have a container that does not exist (ID form):
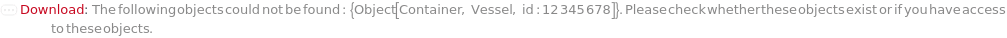
Throw a message if we have a container that does not exist (name form):
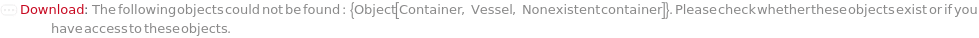
Throw a message if we have a sample that does not exist (ID form):
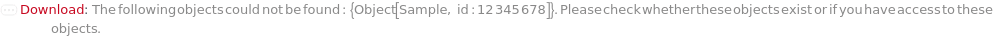
Throw a message if we have a sample that does not exist (name form):
Last modified on Wed 3 Sep 2025 14:58:10