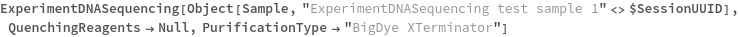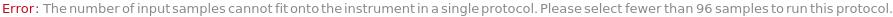ExperimentDNASequencing
ExperimentDNASequencing[Samples]⟹Protocol
creates a Protocol object for running a Sanger DNA sequencing experiment, which uses fluorescently labeled DNA Samples, with a genetic analyzer instrument containing a capillary electrophoresis array to determine the nucleotide sequences from the provided Samples.
ExperimentDNASequencing[Samples,Primers]⟹Protocol
creates a Protocol object for running a Sanger DNA sequencing experiment, which uses fluorescent dideoxynucleotide chain-terminating PCR to label provided Samples using provided Primers and then a genetic analyzer instrument with a capillary electrophoresis array to determine the nucleotide sequences from the provided Samples.
DNA sequencing is the process by which the order of the bases, or nucleotides, in a strand of DNA is determined. Sanger sequencing is a method of DNA sequencing where a DNA sample is reacted with fluorescently labeled chain terminating nucleotides that are incorporated into the DNA strands at different strand lengths. Each fluorophore corresponds to one of the four DNA bases (C, T, G, or A). Once these labeled bases are incorporated, the DNA samples are analyzed using capillary gel electrophoresis (CGE). In CGE, the negatively charged DNA strands are drawn into the polymer-filled capillary via an electrokinetic phenomenon, and the strands separated based on size as they travel through the gel in the capillary. The separated DNA fragments pass a laser that excites the dye molecules. A fluorescence detector measures the fluorescence emission of the dye molecules on the strands. From this emission wavelength, the identity of the base is determined. Sanger sequencing is useful for many applications, including identification of species, confirmation or identification of mutagenesis, or verifying results using next generation sequencing techniques.
Experimental Principles
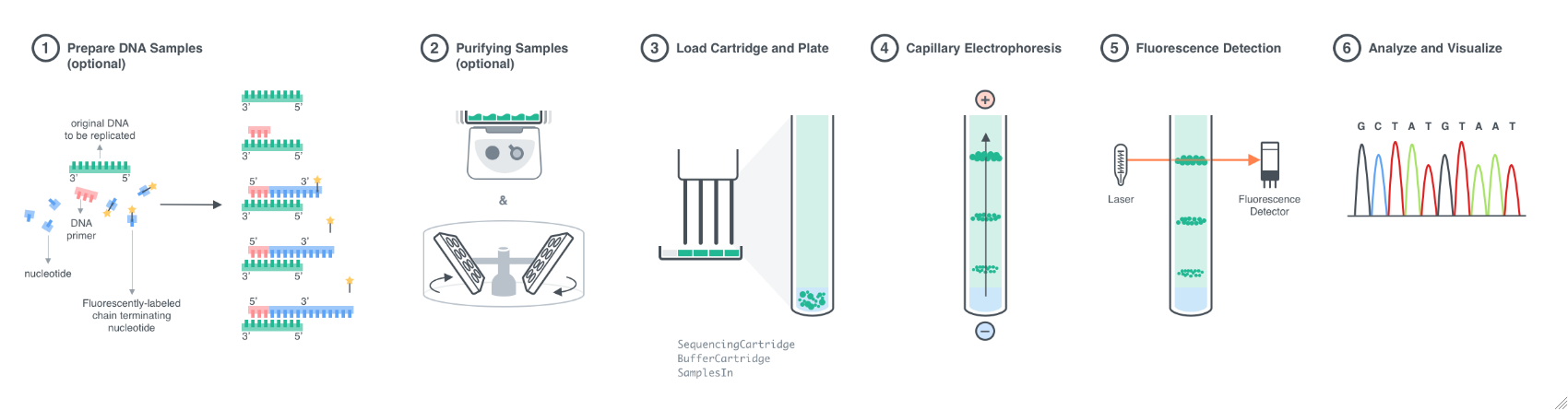
Figure 1.1: Experimental workflow for a DNA sequencing experiment. 1. Prepare samples by loading DNA samples of up to 800 base pairs onto a plate and performing chain termination PCR with fluorescently labeled nucleotides. 2. Purify samples to remove unreacted materials if necessary. 3. Load samples (10-100 Microliter/sample) into autosampler holder of the instrument. The holder can accommodate up to 96 samples in a well plate. Samples must be in groups of 4, as there are 4 capillaries that inject samples from adjacent wells simultaneously; extra wells are filled with buffer. Load the sequencing cartridge containing the capillary array and the buffer cartridge containing the cathode buffer onto the instrument. Start the run. 4. Once the run begins, samples are electrokinetically drawn into the capillaries. Negatively charged DNA fragments travel through the polymer in the capillaries and separate based on size. Longer DNA fragments carry more negative charge compared to shorter fragments. 5. Separated DNA fragments are excited by a laser and fluorescence is detected. 6. Fluorescence data is analyzed using AnalyzeDNASequencing according to calibration to the specific dye set used and the identity of the base in the sequence is determined.
Instrumentation

SeqStudio

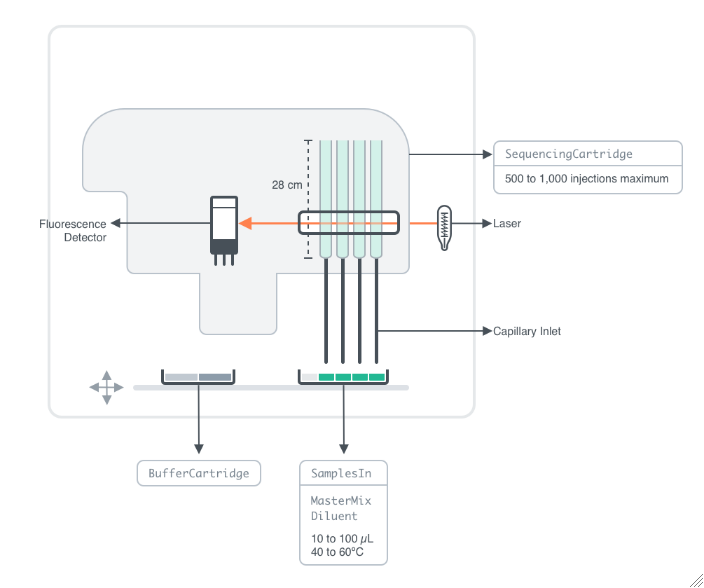
Figure 2.1.1: Instrument schematic of Applied Biosystems SeqStudio Genetic Analyzer, an instrument used for determining the base pair sequence of DNA samples using Sanger sequencing techniques. The SeqStudio comprises a removeable cartridge housing the polymer injection system and capillary array of 4 capillaries, an autosampler, and a fluorescence detector. Once fluorescently labeled samples are loaded and the run method is set up, the instrument electrokinetically draws up samples into the capillaries, and employs capillary gel electrophoresis techniques to separate DNA fragments. The fluorescence detector measures spectral data of emissions from the fluorophores, and further analysis determines the base pairs based on calibration to the fluorophores in the dye set used to label the template. The instrument can analyze up to 96 samples in one run. DNA samples of up to 800 base pairs can be analyzed. Analysis of short DNA strands (<300 base pairs) takes 30 min for 4 samples, processing 96 samples in 12 hours. Analysis of longer strands (>600 base pairs) takes up to 2 hours for 4 samples, and 48 samples can be analyzed in a 24 hour period.
Experiment Options
General
Instrument
The cartridge-based capillary electrophoresis instrument for running the DNA sequencing experiment using fluorescence detection to determine nucleotide sequence.
Pattern Description: An object of type or subtype Model[Instrument, GeneticAnalyzer] or Object[Instrument, GeneticAnalyzer]
Programmatic Pattern: ObjectP[{Model[Instrument, GeneticAnalyzer], Object[Instrument, GeneticAnalyzer]}]
SequencingCartridge
The cartridge containing the polymer, capillary array, and anode buffer that fits into the instrument for running the DNA sequencing experiment.
Pattern Description: An object of type or subtype Object[Item, Cartridge, DNASequencing] or Model[Item, Cartridge, DNASequencing]
Programmatic Pattern: ObjectP[{Object[Item, Cartridge, DNASequencing], Model[Item, Cartridge, DNASequencing]}]
BufferCartridge
The cartridge containing the cathode buffer and waste container that fits into the instrument for running the DNA sequencing experiment.
Pattern Description: An object of type or subtype Object[Container, Vessel, BufferCartridge] or Model[Container, Vessel, BufferCartridge]
Programmatic Pattern: ObjectP[{Object[Container, Vessel, BufferCartridge], Model[Container, Vessel, BufferCartridge]}]
Temperature
Pattern Description: Greater than or equal to 40 degrees Celsius and less than or equal to 60 degrees Celsius.
PreparedPlate
Indicates if the plate containing the samples for the DNA sequencing experiment has been previously prepared with primers and Master Mix and does not need to run sample preparation steps.
Default Calculation: Automatically resolves to False if 'primers' are specified as an input, and True if 'primers' are not an input.
NumberOfInjections
The number of times each amplified sample is injected for a capillary electrophoresis run using identical experimental parameters. All samples are injected for each NumberOfInjections, and a separate Data object is created for each sample for every injection.
Sample Preparation
ReadLength
The estimated number of base pairs in the template samples to be sequenced. Template sample length informs default capillary electrophoresis experiment parameters as described in Figure 3.1.
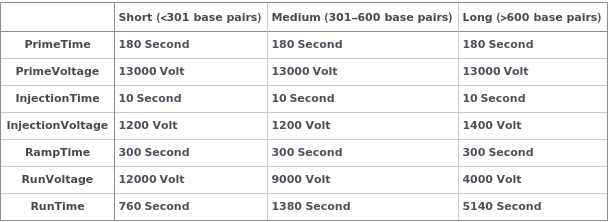
Figure 3.1: Capillary gel electrophoresis separation parameters are set based on the number of base pairs in the sample being analyzed according to the table.
Default Calculation: Automatically resolves based on SequenceLength[Object[Sample][Composition]], or to 500 base pairs if Composition is not known, unless otherwise specified.
SampleVolume
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 100 microliters.
ReactionVolume
The total volume of the polymerase reaction including the template, primer, master mix, and diluent.
Pattern Description: Greater than or equal to 10 microliters and less than or equal to 100 microliters.
PrimerConcentration
Default Calculation: Automatically resolves to 0.1 Micromolar, or Null if PreparedPlate is set to True.
Pattern Description: Greater than or equal to 0.1 picomolar and less than or equal to 10 micromolar or Null.
PrimerVolume
Default Calculation: Automatically set according to the equation PrimerVolume=(PrimerConcentration)*(ReactionVolume)/(inital primer concentration), or 1 Microliter if the calculated volume is too small to be pipetted accurately, or Null otherwise.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 100 microliters or Null.
MasterMix
The stock solution composed of the polymerase, nucleotides, fluorescent dideoxynucleotides, and buffer for the polymerase reaction.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
MasterMixVolume
Default Calculation: Automatically set to 0.5*ReactionVolume if MasterMix is not set to Null, or Null otherwise.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 100 microliters or Null.
AdenosineTriphosphateTerminator
The dye molecule (dideoxynucelotide triphosphate) used to terminate DNA chains at locations where the complementary base on the template strand is thymine.
Default Calculation: Automatically resolves by finding the corresponding dideoxynucleotide dye in the MasterMix Object[Sample][Composition].
Pattern Description: An object of type or subtype Model[Molecule] or Model[ProprietaryFormulation] or Null.
Programmatic Pattern: (ObjectP[{Model[Molecule], Model[ProprietaryFormulation]}] | Automatic) | Null
ThymidineTriphosphateTerminator
The dye molecule (dideoxynucelotide triphosphate) used to terminate DNA chains at locations where the complementary base on the template strand is adenine.
Default Calculation: Automatically resolves by finding the corresponding dideoxynucleotide dye in the MasterMix Object[Sample][Composition].
Pattern Description: An object of type or subtype Model[Molecule] or Model[ProprietaryFormulation] or Null.
Programmatic Pattern: (ObjectP[{Model[Molecule], Model[ProprietaryFormulation]}] | Automatic) | Null
GuanosineTriphosphateTerminator
The dye molecule (dideoxynucelotide triphosphate) used to terminate DNA chains at locations where the complementary base on the template strand is cytosine.
Default Calculation: Automatically resolves by finding the corresponding dideoxynucleotide dye in the MasterMix Object[Sample][Composition].
Pattern Description: An object of type or subtype Model[Molecule] or Model[ProprietaryFormulation] or Null.
Programmatic Pattern: (ObjectP[{Model[Molecule], Model[ProprietaryFormulation]}] | Automatic) | Null
CytosineTriphosphateTerminator
The dye molecule (dideoxynucelotide triphosphate) used to terminate DNA chains at locations where the complementary base on the template strand is guanine.
Default Calculation: Automatically resolves by finding the corresponding dideoxynucleotide dye in the MasterMix Object[Sample][Composition].
Pattern Description: An object of type or subtype Model[Molecule] or Model[ProprietaryFormulation] or Null.
Programmatic Pattern: (ObjectP[{Model[Molecule], Model[ProprietaryFormulation]}] | Automatic) | Null
Diluent
The solution for bringing each reaction to ReactionVolume once all the reaction components (template, primers, and master mix) are added.
Default Calculation: Automatically resolves to Model[Sample,"Nuclease-free Water"], or Null if PreparedPlate is set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
DiluentVolume
Default Calculation: Automatically set according to the equation DiluentVolume=ReactionVolume-(SampleVolume+MasterMixVolume+PrimerVolume) if Diluent is not set to Null, or Null otherwise.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 100 microliters or Null.
Post Experiment
PrimerStorageCondition
For each sample, the non-default conditions under which the primers of this experiment should be stored after the protocol is completed. If left unset, the primers will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
MasterMixStorageCondition
For each sample, the non-default condition under which MasterMix of this experiment should be stored after the protocol is completed. If left unset, MasterMix will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Polymerase Activation
Activation
Indicates if hot start activation should be performed in the polymerase reaction. In order to reduce non-specific amplification, enzymes can be made room temperature stable by inhibiting their activity via thermolabile conjugates. Once an experiment is ready to be run, this inhibition is disabled by heating the reaction to ActivationTemperature.
Default Calculation: Automatically set to True if other Activation options are set, or False otherwise.
ActivationTime
The length of time for which the sample is held at ActivationTemperature to remove the thermolabile conjugates inhibiting polymerase activity in the polymerase reaction.
Default Calculation: Automatically set to 60 seconds if Activation is set to True, or Null otherwise.
ActivationTemperature
The temperature to which the sample is heated to remove the thermolabile conjugates inhibiting polymerase activity in the polymerase reaction.
Default Calculation: Automatically set to 95 degrees Celsius if Activation is set to True, or Null otherwise.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 105 degrees Celsius or Null.
ActivationRampRate
Default Calculation: Automatically set to 3.5 degrees Celsius per second if Activation is set to True, or Null otherwise.
Pattern Description: Greater than or equal to 0.1 degrees Celsius per second and less than or equal to 3.5 degrees Celsius per second or Null.
Denaturation
DenaturationTime
The length of time for which the sample is held at DenaturationTemperature to allow the dissociation of the double stranded template into single strands in the polymerase reaction.
DenaturationTemperature
The temperature to which the sample is heated to allow the dissociation of the double stranded template into single strands in the polymerase reaction.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 105 degrees Celsius or Null.
DenaturationRampRate
Pattern Description: Greater than or equal to 0.1 degrees Celsius per second and less than or equal to 3.5 degrees Celsius per second or Null.
Primer Annealing
PrimerAnnealing
Indicates if annealing should be performed as a separate step instead of as part of extension in the polymerase reaction. Lowering the temperature during annealing allows primers to bind to the template and serve as anchor points for the polymerase in the subsequent extension.
Default Calculation: Automatically set to True if other PrimerAnnealing options are set, or False otherwise.
PrimerAnnealingTime
The length of time for which the sample is held at PrimerAnnealingTemperature to allow primers to bind to the template in the polymerase reaction.
Default Calculation: Automatically set to 5 seconds if PrimerAnnealing is set to True, or Null otherwise.
PrimerAnnealingTemperature
The temperature to which the sample is cooled to allow primers to bind to the template in the polymerase reaction.
Default Calculation: Automatically set to 50 degrees Celsius if PrimerAnnealing is set to True, or Null otherwise.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 105 degrees Celsius or Null.
PrimerAnnealingRampRate
The rate at which the sample is cooled to reach PrimerAnnealingTemperature in the polymerase reaction.
Default Calculation: Automatically set to 1 degrees Celsius per second if PrimerAnnealing is set to True, or Null otherwise.
Pattern Description: Greater than or equal to 0.1 degrees Celsius per second and less than or equal to 3.5 degrees Celsius per second or Null.
Strand Extension
ExtensionTime
The length of time for which sample is held at ExtensionTemperature to allow the polymerase to synthesize a new strand using the template and primers in the polymerase reaction.
ExtensionTemperature
The temperature to which the sample is heated/cooled to allow the polymerase to synthesize a new strand using the template and primers in the polymerase reaction.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 105 degrees Celsius or Null.
ExtensionRampRate
The rate at which the sample is heated/cooled to reach ExtensionTemperature in the polymerase reaction.
Pattern Description: Greater than or equal to 0.1 degrees Celsius per second and less than or equal to 3.5 degrees Celsius per second or Null.
Thermocycling
NumberOfCycles
The number of times the polymerase reaction will undergo repeated cycles of denaturation, primer annealing (optional), and strand extension.
Pattern Description: Greater than or equal to 1 and less than or equal to 60 in increments of 1 or Null.
HoldTemperature
The temperature to which the sample is cooled and held after the polymerase reaction thermocycling procedure.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 105 degrees Celsius or Null.
Post-PCR Sample Preparation
PurificationType
The method of purification of the DNA template samples after undergoing chain termination PCR. Ethanol precipitation selectively precipitates DNA out of solution by adding solutions of ethanol and EDTA to the sample, mixing, and centrifuging. Then the supernatant containing reaction reagents is discarded and the pure DNA pellet is resuspended. BigDye XTerminator purification removes unincorporated BigDye terminators and salts by adding BigDye XTerminator solution to the reactions, then mixing and centrifuging.
Default Calculation: Automatically set to Null if PreparedPlate->True, or BigDye XTerminator if a BigDye master mix is used, or Ethanol precipitation otherwise.
QuenchingReagents
Default Calculation: Automatically set to Model[Sample,"Absolute Ethanol, Anhydrous"] and Model[Sample, StockSolution, "125 mM EDTA"] if PurificationType->Ethanol precipitation, or Model[Sample,"SAM Solution"] and Model[Sample,"BigDye XTerminator Solution"] if PurificationType->BigDye XTerminator, or Null otherwise.
Pattern Description: List of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample entries or Null.
QuenchingReagentVolumes
Default Calculation: Automatically set to {30 Microliter, 1 Microliter} if PurificationType->Ethanol precipitation, or {4.5*ReactionVolume, ReactionVolume} of the ReactionVolume of the first sample if PurificationType->BigDye XTerminator, or Null otherwise.
Pattern Description: List of one or more greater than or equal to 1 microliter and less than or equal to 100 microliters entries or Null.
SequencingBuffer
The buffer to be used to resuspend DNA samples prior to loading in the genetic analyzer instrument. This buffer will also be used to fill empty wells that are needed to complete injection groups as a blank.
Default Calculation: Automatically set to Model[Sample,"Tris EDTA 0.1 buffer solution"] if PurificationType->Ethanol precipitation, or Null if PurificationType->BigDye XTerminator and otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
SequencingBufferVolume
The volume of the buffer to be used to resuspend DNA samples prior to loading in the genetic analyzer instrument.
Default Calculation: Automatically set to 40 microliters if PurificationType->Ethanol precipitation, or Null if PurificationType->BigDye XTerminator and otherwise.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 100 microliters or Null.
Separation
DyeSet
The set of dye terminator molecules used to terminate DNA chains, with different emission spectra corresponding to the different nucleotides. For optimal performance use the dye set that most closely matches the Master Mix and dyes used in the polymerase reaction.
Programmatic Pattern: ("E_BigDye Terminator v1.1" | "Z_BigDye Terminator v3.1" | "Z_BigDye Direct") | Automatic
PrimeTime
The length of time for which cathode buffer is drawn into the capillary array in order to prime the capillaries prior to the samples being injected.
PrimeVoltage
The voltage applied to the capillary array in order to prime the capillaries prior to the samples being injected.
InjectionTime
The length of time for which sample is drawn into the capillary array. A longer injection time will lead to an increase in the signal, as more DNA molecules will be electrokinetically drawn into the capillary.
InjectionVoltage
Default Calculation: Automatically set according to the ReadLength option specifying the length of the sequence to be read.
RampTime
RunVoltage
The voltage applied to the capillary array while the template samples move through the capillary and the fragments separate.
Default Calculation: Automatically set according to the ReadLength option specifying the length of the sequence to be read.
RunTime
The length of time for which the separation of the template fragments in the capillaries will occur.
Default Calculation: Automatically set according to the ReadLength option specifying the length of the sequence to be read.
Storage
SequencingCartridgeStorageCondition
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
PreparatoryPrimitives
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more a primitive with head Define, Transfer, Mix, Aliquot, Consolidation, FillToVolume, Incubate, Filter, Wait, Centrifuge, or Resuspend entries or Null.
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentrationFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Basics
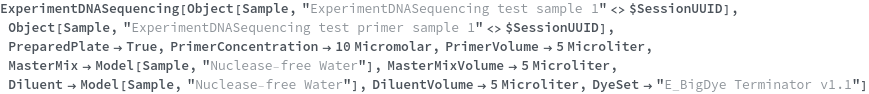
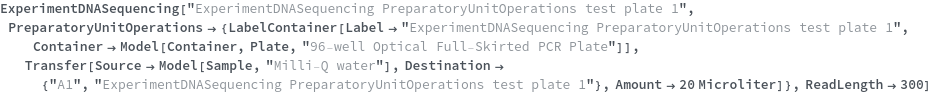
To perform a DNA sequencing experiment on samples that are already fluorescently labeled, simply run:

Primers may be specified to indicate that they should be added to the reaction mixtures and that chain termination PCR wil be used to prepare the samples:

PreparedPlate
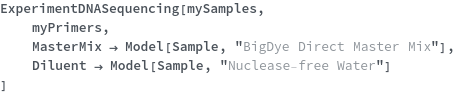
If samples will undergo chain termination PCR, add master mix containing dyes, nucleotides and buffer, as well as a diluent to prepare the samples for capillary electrophoresis
Set PCR settings
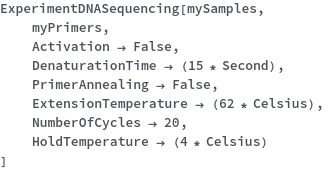
If a PreparedPlate is not being used, settings for the chain termination polymerase reaction can be set
PurificationType

DyeSet

Capillary Electrophoresis
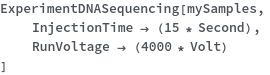
Warnings and Errors
Messages (36)
DeprecatedModels (2)
DiscardedSamples (2)
DNASequencingAnodeBufferInvalid (1)
DNASequencingBufferVolumeMismatch (2)
DNASequencingCartridgeStorageCondition (1)
DNASequencingCartridgeTypeInvalid (1)
DNASequencingCathodeBufferInvalid (1)
DNASequencingDiluentNotSpecified (1)
DNASequencingMasterMixNotSpecified (2)
DNASequencingMasterMixStorageConditionMismatch (1)
DNASequencingPCROptionsForPreparedPlate (1)
DNASequencingPolymerInvalid (1)
DNASequencingPreparedPlateInputInvalid (1)
DNASequencingPreparedPlateMismatch (2)
DNASequencingPrimerCompositionNull (1)
DNASequencingPrimerStorageConditionMismatch (1)
DNASequencingPurificationTypeMismatch (2)
DNASequencingQuenchingReagentVolumeMismatch (2)
DNASequencingReadLengthNotSpecified (1)
DNASequencingTooManyInjectionGroups (1)
DNASequencingTooManySamples (1)
DuplicateName (1)
InvalidPreparedPlateContainer (2)
MultiplePrimerSampleOligomersSpecified (1)
MultipleSampleOligomersSpecified (1)
Possible Issues
Too much or too little DNA template
The amount of DNA in the sample is important, and should be quantified before preparing samples for DNA sequencing. Based on the length of the DNA sample, the concentration of the DNA samples should be changed. An amount of 20 nanograms of PCR product in a 10 microliter reaction is the minimum. More DNA should be used for longer sequences.

Dye Blobs
Indicates poor removal of the dye terminators. If BigDye products were used, purification with BigDye XTerminator is recommended to remove unreacted dyes prior to capillary electrphoresis.

Off scale peaks
If the amplicon is very efficient, or if too much DNA is in the sample, off scale peaks can occur. To fix this, adjust the InjectionTime or InjectionVoltage options, or use a lower concentration of DNA in the sample, or reduce the number of cycles in the PCR experiment to prepare the samples for ExperimentDNASequencing.

Last modified on Thu 29 Dec 2022 10:45:41