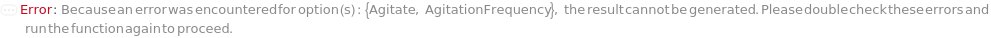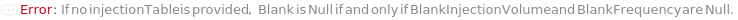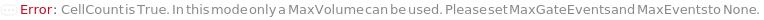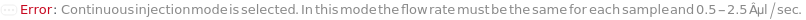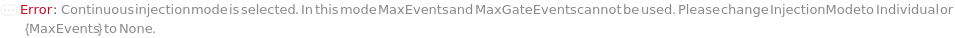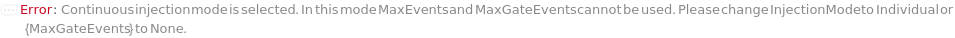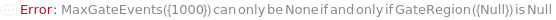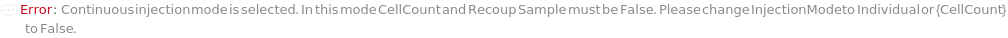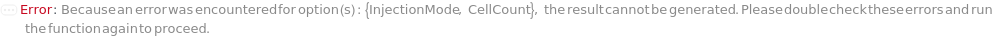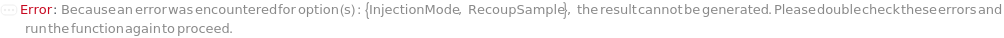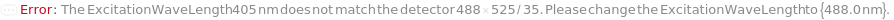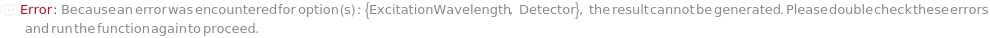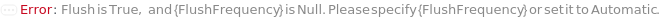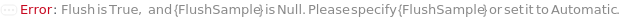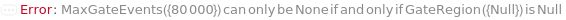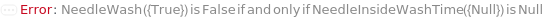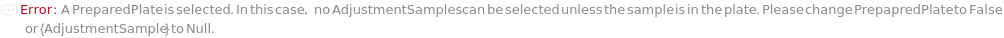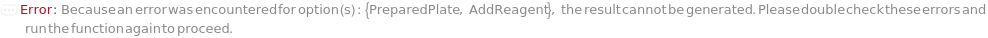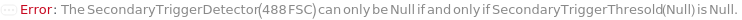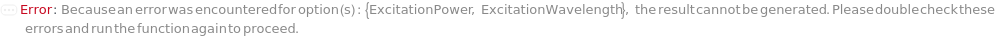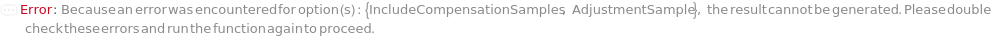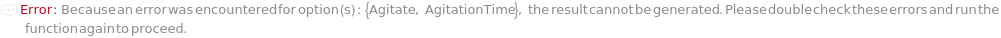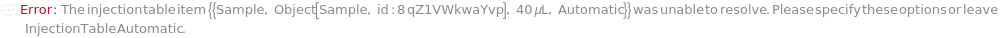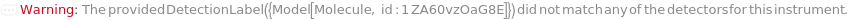ExperimentFlowCytometry
ExperimentFlowCytometry[Samples]⟹Protocol
generates a Protocol to anaylze Samples via flow cytometry.
Flow cytometry is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles flowing single file in a stream of fluid. The single-cell suspension creates unique light-scattering events that occur when each cell passes through laser light. The scattered light is characteristic of the size and internal complexity of the cell, as well as the intensity of the signal that is generated by the specific dyes, thus creating patterns that reflect cell type. This approach makes flow cytometry a powerful tool for detailed analysis of complex populations in a short period of time.
Experimental Principles
Figure 1.1: Procedural overview of a flow cytometry experiment. Step 1: Plate is loaded with sample. Step 2: Quality control beads are run through the instrument. Step 3: The PMT voltages are adjusted using each detectors adjustment sample (if specified). Step 4: The adjustment samples are run through the instrument to determine the compensation matrix (if specified). Step 4: The samples are run through the instrument. Step 5: The leftover sample in the injection tubing is backflushed back into the sample (optional).
Instrumentation
ZE5 Cell Analyzer
Figure 2.1: Instrument diagram for the ZE5 Flow Cytometer. Once the sheath fluid is running at laminar flow, the cells are injected into the center of the stream, at a slightly higher pressure. The principles of hydrodynamic focusing cause the cells to align, single file in the direction of flow. Water is used as a sheath fluid with a small amount surfactant added, helping keep the system flowing by reducing the surface tension. The sample flows by 5 different spatially separated laser and detector sets. The Forward Scatter detects elastically scattered light 2-18 degrees off the axis of the laser and is proportional to the perimeter of the cross section of the cell. The Side Scatter detects elastically scattered light 90 degrees off the axis of the laser and indicates the relative differences in particle complexity (granularity, membrane structure, and cytoplasmic constituents). The Fluorescent detectors measure light that fluorophores emit absorbing the photons from the excitation laser. The emitted light passes through a series filters to only detect light in a specific wavelength band associated with the emission wavelengths specific fluorophores. The filters used by this instrument are: Dichroic Short Pass (DSP, wavelengths smaller than the threshold pass through and large wavelengths are reflected), Dichroic Long Pass (DLP, wavelengths larger than the threshold pass through and small wavelengths are reflected), Long Pass (LP, wavelengths larger than the threshold pass through and small wavelengths filtered out), Band Pass (passes wavelengths within a certain band and filters out other wavelengths), Notch (filters out wavelengths within a certain band and lets other wavelengths pass) and Neutral Density (ND, reduces the intensity of all wavelengths by the filters optical density (OD), the transmitted light intensity is 10^(-OD) the intensity of the incident light.)
Experiment Options
General
InjectionMode
The mode in which samples are loaded into the flow cell. In the Individual injection mode, only one sample is inside the line a given time. After data acquisition, the sample pump runs backward to clear the line, then a wash occurs before the probe moves to the next position. The sample can be returned to the originional container if desired. In Continuous mode, samples are aspirated continuously, resulting multiple samples in the sample line. Each sample is separated with a series of air and water boundaries. In this mode, samples cannot be returned and reagent cannot be added to samples.
RecoupSample
Indicates if the excess sample in the injection line returns to the into the container that they were in prior to the measurement.
Default Calculation: Automatically set to True for Individual InjectionMode and Null for Continuous InjectionMode.
SampleVolume
Default Calculation: Is automatically set to 200 Microliter if MaxVolume is less than 200Microliter for all samples, otheriwise it is set to twice the MaxVolume up to 4Milliliter. If PreparedPlate is True it is set to Null.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 4 milliliters or Null.
Instrument
The flow cytometer used for this experiment to count and quantify the cell populations by aligning them in a stream and measuring light scattered off the cells.
Pattern Description: An object of type or subtype Object[Instrument, FlowCytometer] or Model[Instrument, FlowCytometer]
Programmatic Pattern: ObjectP[{Object[Instrument, FlowCytometer], Model[Instrument, FlowCytometer]}]
FlowCytometryMethod
The flow cytometry method object which describes the optics settings of the instrument. Specific parameters of the object can be overridden by other flow cytometry options.
PreparedPlate
Indicates if the container containing the samples for the flow cytometry experiment has been previously prepared and will be loaded directly onto the instrument.
Temperature
The temperature of the autosampler where the samples sit prior to being injected into the flow cytometer.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 37 degrees Celsius in increments of 1 degree Celsius or Null.
Agitate
Indicates if the autosampler is shaked to the resuspend sample prior to being injected into the flow cytometer.
AgitationFrequency
The frequency at which the autosampler is shaked to resuspend samples between the injections (with a value of 2 indicating agitating after every 2 samples or First indicating to agitate before the first sample).
AgitationTime
The amount of time the autosampler is shaked to the resuspend sample prior to being injected into the flow cytometer.
FlowRate
The volume or number of trigger events per time (see diagram) in which the sample is flowed through the flow cytometer.
Pattern Description: Greater than or equal to 0.1 microliters per second and less than or equal to 3.5 microliters per second or greater than or equal to 0 Events per second and less than or equal to 100000 Events per second or Null.
Programmatic Pattern: (RangeP[0.1*(Microliter/Second), 3.5*(Microliter/Second)] | RangeP[0*(Event/Second), 100000*(Event/Second)]) | Null
CellCount
Indicates if the number of cells per volume of the sample will be measured. This setting an adddional 5ul is run before aquisition to stabilize flow.
NumberOfReplicates
Pattern Description: Greater than or equal to 0 and less than or equal to 384 in increments of 1 or Null.
Flushing
Flush
Indicates if a sample line flush is performed after the FlushFrequency number of samples have been processed.
FlushFrequency
Indicates the frequency at which the flushed after samples have been processed (with a value of 2 indicating flushing after every 2 samples).
FlushSample
The liquid used to flush the instrument between FlushFrequency number of samples to clean the sample line.
Default Calculation: Automatically set to Model[Sample,StockSolution"Filtered PBS, Sterile"] if Flush is True; otherwise, set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
FlushSpeed
Default Calculation: Automatically set to 0.5Microliter/Second if Flush is True; otherwise, set to Null.
Pattern Description: 0.5 microliters per second, 1 microliter per second, or 1.5 microliters per second or Null.
Programmatic Pattern: ((0.5*(Microliter/Second) | 1*(Microliter/Second) | 1.5*(Microliter/Second)) | Automatic) | Null
Optics
Detector
Default Calculation: Automatically set to 488 FSC (forward scatter), 488 SSC (side scatter) and the optic modules whose excitation wavelengths and detection wavelengths closet match the excitation and emission spectra of the DetectionLabels. If no DetectionLabel is found 488 525/35 is selected.
Pattern Description: 488 FSC, 405 FSC, 488 SSC, 488 525/35, 488 593/52, 488 750LP, 488 692/80, 561 750LP, 561 670/30, 561 720/60, 561 589/15, 561 577/15, 561 640/20, 561 615/24, 405 670/30, 405 720/60, 405 750LP, 405 460/22, 405 420/10, 405 615/24, 405 525/50, 355 525/50, 355 670/30, 355 700LP, 355 447/60, 355 387/11, 640 720/60, 640 775/50, 640 800LP, or 640 670/30.
DetectionLabel
Indicates the fluorescent tags, attached to the samples that will be analyzed. This is used to automatically determine the Detector used.
Default Calculation: Automatically set to the model of the fluorophores in the composition of the sample.
NeutralDensityFilter
Indicates if a neutral density filter with an optical density of 2.0 should be used lower the intensity of scattered light that hits the detector. This will decrease the intensity of the scattered light by blah to reduce the signal to the PMTs dynamic range. This is only applicable for forward scatter detectors.
Default Calculation: Automatically set to the Gain field in the FlowCytometryMethod if provided and False otherwise.
Gain
The voltage the PhotoMultiplier Tube (PMT) should be set to to detect the scattered light off the sample. When QualityControl is selected, the PMT voltage values are those set at the Quality Control baseline run at the begining of the experiment. When Auto is selected, PMT voltage is optimized at the begining of the experiment. The gain value is set so that the intensity signal from the AdjustmentSample will reach the TargetSaturationPercentage of the PMTs dynamic range. The gain optimization is performed by measuring the AdjustmentSample and increasing or decreasing the gain value until the TargetSaturationPercentage is reached.
Default Calculation: Automatically set to the Gain field in the FlowCytometryMethod if provided and QC otherwise.
Pattern Description: Auto or QualityControl or greater than or equal to 0 volts and less than or equal to 1000 volts.
AdjustmentSample
When optimizing gain, the sample used for gain optimization. If IncludeCompensationSamples is True, this sample will also be used when creating a compensation matrix.
Default Calculation: Automatically set to a sample with the DetectionLabel in the sample's composition if Gain are Auto.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
TargetSaturationPercentage
When optimizing gain, the gain is set so that the intensity of positive population is centered around the 'TargetSaturationPercentage' of the PMT's dynamic range.
Default Calculation: Automatically set to 75 Percent if any of the gain optimization parameters are specified.
Pattern Description: Greater than or equal to 1 percent and less than or equal to 99 percent or Null.
ExcitationWavelength
Pattern Description: 355 nanometers, 405 nanometers, 488 nanometers, 561 nanometers, or 640 nanometers.
ExcitationPower
Pattern Description: Greater than or equal to 0 milliwatts and less than or equal to 100 milliwatts.
Trigger
TriggerDetector
The detector used to determine what signals count as an event. The TriggerDetector, combined with the TriggerThreshold, defines real events that should be detected and analyzed. Typically, a forward scatter detector is selected for the trigger because it identifies all particles above a given size regardless of the fluorescent signal.
Pattern Description: 488 FSC, 405 FSC, 488 SSC, 488 525/35, 488 593/52, 488 750LP, 488 692/80, 561 750LP, 561 670/30, 561 720/60, 561 589/15, 561 577/15, 561 640/20, 561 615/24, 405 670/30, 405 720/60, 405 750LP, 405 460/22, 405 420/10, 405 615/24, 405 525/50, 355 525/50, 355 670/30, 355 700LP, 355 447/60, 355 387/11, 640 720/60, 640 775/50, 640 800LP, or 640 670/30.
TriggerThreshold
The level of the intensity detected by TriggerDetector must fall above to be classified as an event. The value is reported as a percentage of the total range of signal intensities in that detector.
SecondaryTriggerDetector
The additional detector used to determine what signals count as an event. The SecondaryTriggerDetector, combined with the SecondaryTriggerThreshold, defines real events that should be detected and analyzed.
Pattern Description: {488 FSC, 405 FSC, 488 SSC, 488 525/35, 488 593/52, 488 750LP, 488 692/80, 561 750LP, 561 670/30, 561 720/60, 561 589/15, 561 577/15, 561 640/20, 561 615/24, 405 670/30, 405 720/60, 405 750LP, 405 460/22, 405 420/10, 405 615/24, 405 525/50, 355 525/50, 355 670/30, 355 700LP, 355 447/60, 355 387/11, 640 720/60, 640 775/50, 640 800LP, 640 670/30} or None or Null.
SecondaryTriggerThreshold
The level of the intensity detected by SecondaryTriggerDetector must fall above to be classified as an event. The value is reported as a percentage of the total range of signal intensities in that detector.
Default Calculation: Automatically set to 10 Percent if the SecondaryTriggerDetector is not None; otherwise, set to Null.
Pattern Description: Greater than or equal to 0.01 percent and less than or equal to 100 percent or Null.
Stopping Conditions
MaxVolume
The maximum volume of each sample that will flow through the flow cytometer before it moves on to the next sample. After this volume, the acquisition for this sample will stop and the next sample will begin. If the MaxEvents and/or MaxGateEvents are set, the acquisition will end once it the first maxima is reached.
Pattern Description: Greater than or equal to 10 microliters and less than or equal to 4000 microliters.
MaxEvents
The maximum number of trigger events that will flow through the flow cytometer. The acquisition for this sample will stop and the next sample will begin once it the first maxima (MaxVolume, MaxEvents or MaxGateEvents) is reached.
MaxGateEvents
The maximum events falling into a specific Gate that will flow through the instrument. The acquisition for this sample will stop and the next sample will begin once it the first maxima (MaxVolume, MaxEvents or MaxGateEvents) is reached.
GateRegion
The conditions given to categorize the gate for the MaxGateEvents. Events that fall within the X Range and Y Range of the X Channel (area) by Y Channel (area) plot will count towards the MaxGateEvents.
Programmatic Pattern: {ListableP[FlowCytometryDetectorP], RangeP[0*Percent, 100*Percent] ;; RangeP[0*Percent, 100*Percent], ListableP[FlowCytometryDetectorP], RangeP[0*Percent, 100*Percent] ;; RangeP[0*Percent, 100*Percent]} | Null
Compensation
IncludeCompensationSamples
Indicates if a compensation matrix will be created to compensate for spillover of DetectionLabel into other detectors. The AdjustmentSample for each fluorescent detector will be measured to calculate this matrix.
UnstainedSample
A unstained sample to be used as negative control when calculating the background of the compensation matrix for the experiment. This is needed if any of the AdjustmentSamples do not have negative populations.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or None or Null.
Reagent Addition
AddReagent
Reagent
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ReagentVolume
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 5000 microliters or Null.
ReagentMix
Washing
NeedleWash
Indicates if sheath fluid will be used to wash the injection needle after the sample measurement. The outside of the needle is washed for 0.25 Second after every sample measurement.
NeedleInsideWashTime
InjectionOrder
BlankFrequency
The frequency at which Blank samples will be inserted in the measurement sequence. If specified to a number, indicates how often the Blank samples will run among the samples; for example, if 5 input samples are measured and BlankFrequency is 2, then Blank measurement will occur after the first two samples and again after the fourth.
Blank
A sample containing wash solution used an extra wash between samples. The blank will flow through the instrument like a sample without detecting scattering.
Default Calculation: Automatically set to Model[Sample,StockSolution"Filtered PBS, Sterile"] if any other Blank option is selected; otherwise, set to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BlankInjectionVolume
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 2000 microliters or Null.
InjectionTable
The order of Sample, AdjustmentSample, UnstainedSample and Blank sample loading into the flow cytometer.
Default Calculation: Determined to the order of input samples articulated. AdjustmentSamples and UnstainedSample are inserted at the beginning of the experiment followed by samples. The Blank samples are inserted based on the BlankFrequency.
Programmatic Pattern: {{Sample | AdjustmentSample | UnstainedSample | Blank, Alternatives[ObjectP[{Model[Sample], Object[Sample]}] | _String], RangeP[0*Microliter, 4*Milliliter] | Automatic, RangeP[5*Second, 1*Hour] | Automatic | 0*Second}..} | Automatic
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSamplePreparation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentratedBufferDilutionFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Agitation
Flush
Detectors
Specify the detectors which should be used to detect light scattered off the samples and the fluorophore molecule the detector is meant to excite:
Gain
Specify the voltage the PhotoMultiplier Tube (PMT) should be set to to detect the scattered light off the sample. When QualityControl is selected, the PMT voltage values are those set at the Quality Control baseline run at the begining of the experiment. When Auto is selected, PMT voltage is optimized at the begining of the experiment. The gain value is set so that the intensity signal from the AdjustmentSample will reach the TargetSaturationPercentage of the PMTs dynamic range:
Trigger
Stopping Conditions
Specify the maximum volume and number of trigger events that will flow through the flow cytometer. The acquisition for this sample will stop and the next sample will begin once it the first maxima is reached:
Compensation
Specify the compensation samples that should be included to make a compensation table after the experiment. Include an unstained sample if the samples do not have negative populations:
Reagent Addition
Warnings and Errors
Messages (40)
ConflictingAddReagentOptions (1)
ConflictingBlankOptions (1)
ConflictingContinuousModeFlowRateOptions (2)
ConflictingContinuousModeLimitOptions (2)
ConflictingContinuousModeOptions (2)
ConflictingFlowInjectionTable (1)
ConflictingFlowInjectionTableBlankOptions (1)
ConflictingFlushInjectionOptions (1)
ConflictingFlushOptions (2)
ConflictingPreparedPlateAdjustmentSampleOptions (1)
ConflictingPreparedPlateBlankOptions (1)
ConflictingPreparedPlateInjectionOrder (1)
ConflictingPreparedPlateReagentOptions (1)
ConflictingPreparedPlateUnstainedSampleOptions (1)
ConflictingSecondaryTriggerOptions (1)
FlowCytometryInvalidPreparedPlateContainer (2)
FlowCytometryTooManySamples (1)
GainOptimizationOptions (1)
MustSpecifyAdjustmentSamplesForCompensation (1)
ObjectDoesNotExist (6)
Do NOT throw a message if we have a simulated container but a simulation is specified that indicates that it is simulated:
Do NOT throw a message if we have a simulated sample but a simulation is specified that indicates that it is simulated:
Throw a message if we have a container that does not exist (ID form):
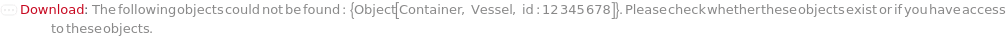
Throw a message if we have a container that does not exist (name form):
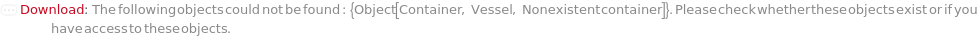
Throw a message if we have a sample that does not exist (ID form):
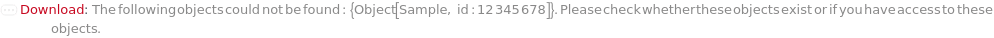
Throw a message if we have a sample that does not exist (name form):
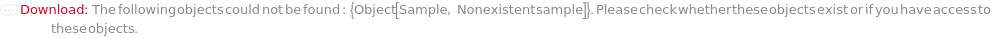
UnresolvableFlowCytometerInjectionTable (1)
Last modified on Tue 2 Sep 2025 18:20:27