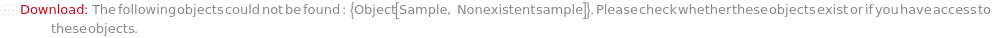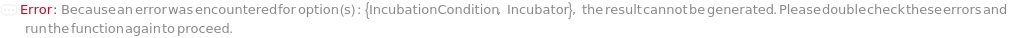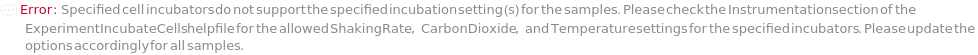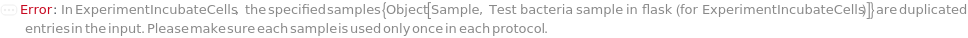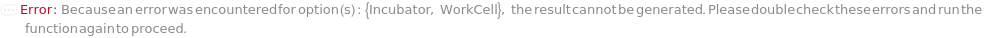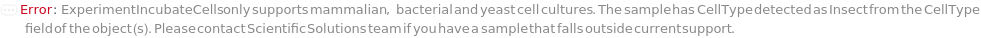ExperimentIncubateCells
ExperimentIncubateCells[Samples]⟹Protocol
creates a Protocol object to grow or maintain the provided Samples containing mammalian, yeast, or bacterial cells, with desired incubation conditions.
Experimental Principles
Instrumentation
Default Cell Incubators
Custom Cell Incubators
Cytomat HERAcell 240i TT 10
STX44-ICBT with Humidity Control
Bactomat HERAcell 240i TT 10 for Yeast
Bactomat HERAcell 240i TT 10 for Bacteria
STX44-ICBT with Shaking
Innova 44 for Bacterial Plates
Innova 44 for Bacterial Flasks
Innova 44 for Yeast Plates
Innova 44 for Yeast Flasks
Multitron Pro with 3mm Orbit
Multitron Pro with 25mm Orbit
Experiment Options
General
Preparation
WorkCell
Time
Incubator
Incubation Condition
Temperature
Shake
ShakingRate
ShakingRadius
IncubationCondition
CellType
CultureAdhesion
RelativeHumidity
CarbonDioxide
Post Experiment
SamplesOutStorageCondition
Protocol Options
Organizational Information
Template
Name
Example Results
- Here we are enqueuing a protocol to start an E. coli liquid culture. This protocol performs a manual workflow that inoculates from an E. coli AgarStab stock into a flask containing LB broth, incubates the flask in a bacterial incubator with fully customized parameters, and quantifies the resulting cell density.
- Inspecting the actual environmental data recorded during cell incubation in the custom incubator, which is equipped with sensors for CarbonDioxide, RelativeHumidity, and Temperature.
- The same data is also collectively stored in the IncubatorEnvironmentalData field of the subprotocol
- Here, the sample appears as both SamplesIn and SamplesOut because it is the same sample and container before and after incubation.
- Inspecting the cell density change of the incubated sample by comparing the Model[Cell] entry before the QuantifyCells unit operation and after completion of the entire protocol.
- At the beginning of the IncubateCells subprotocol, the sample’s Composition contains a Model[Cell] entry with the cell concentration estimated based on the amount transferred to the flask during inoculation from the agar stab
- We got an OD600 update! After the IncubateCells unit operation finishes, the QuantifyCells unit operation measures the concentration of Model[Cell] and updates the Composition of the incubated samples.
- The OD600 measurement data is also stored as an object in both the Object[Sample] and the QuantifyCells subprotocol
- Watching the video streams of the InoculateLiquidMedia subprotocol performed in the biosafety cabinet prior to the IncubateCells subprotocol
- Here we are enqueuing a script to start an E. coli culture on agar plates from freeze-dried powder. This script involves a Robotic unit operation that spreads the resuspended and serial-diluted E Coli cell suspension onto several agar plates, a Manual unit operation that incubates some of the most-diluted agar plates in a shared non-shaking bacterial incubator, and finally another Robotic unit operation that quantifies the colonies grown.
- Checking what protocols are generated by this script run, along with their OutputUnitOperations and Subprotocols
- Extracting the IncubateCells subprotocol, which is the only subprotocol of the second ManualCellPreparation protocol in the script. We will be looking more closely at this subprotocol.
- Inspecting the actual environmental data during cell incubation in this shared bacterial incubator, which is equipped with Temperature and RelativeHumidity sensors.
- Extracting the data from individual environmental data fields of the IncubateCells subprotocol. Each field contains data whose lengths match the number of input agar plate samples.
- The same data is also collectively stored in the IncubatorEnvironmentalData field of the subprotocol
- Visualizing these sensor data, we observed some fluctuations in both Temperature and RelativeHumidty, likely caused by opening/closing the door of the incubator.
customFlaskProtocol = ExperimentCellPreparation[
{
InoculateLiquidMedia[
Sample -> Model[Sample, "E.coli DH5 Alpha Agar Stab"],
DestinationMedia -> Model[Sample, Media, "LB (Liquid)"],
DestinationMediaContainer -> Model[Container, Vessel, "250mL Erlenmeyer Flask"],
ContainerOutLabel -> "My Flask of Inoculated Cells"
],
IncubateCells[
Sample -> "My Flask of Inoculated Cells",
IncubationCondition -> Custom,
CellType -> Bacterial,
CultureAdhesion -> Suspension,
Shake -> True,
ShakingRate -> 100 RPM,
Temperature -> 35 Celsius,
CarbonDioxide -> 7 Percent,
RelativeHumidity -> 80 Percent,
Time -> 24 Hour
],
QuantifyCells[
Sample -> "My Flask of Inoculated Cells",
SamplesInStorageCondition -> Refrigerator
]
}
]
bacterialIncubationPlateScript = ExperimentCellPreparation[
{
LabelSample[
Sample -> Model[Sample, "E.coli 10798"],
Amount -> 1.5 Gram,
ExactAmount -> False,
Container -> Model[Container, Vessel, "5mL small glass vial aluminum cap"],
Label -> "my Ecoli powder"
],
SpreadCells[
Sample -> "my Ecoli powder",
InoculationSource -> FreezeDried,
ResuspensionMedia -> Model[Sample, Media, "LB (Liquid)"],
DilutionType -> Serial,
Diluent -> Model[Sample, Media, "LB (Liquid)"],
NumberOfDilutions -> 4,
SerialDilutionFactor -> 10,
ContainerOutLabel -> {"My container 1", "My container 2", "My container 3", "My container 4", "My container 5"}
],
IncubateCells[
Sample -> {"My container 4", "My container 5"},
IncubationCondition -> BacterialIncubation,
CellType -> Bacterial,
CultureAdhesion -> SolidMedia,
Shake -> False,
Time -> 12 Hour
],
QuantifyColonies[
Sample -> {"My container 4", "My container 5"}
]
}
]
Warnings and Errors
Messages (76)
CellTypeNotSpecified (1)
ConflictingCellType (2)
ConflictingCellTypeWithCultureAdhesion (2)
ConflictingCellTypeWithIncubationCondition (2)
If CellType for samples and SamplesOutStorageCondition are both microbial but one is Bacterial and the other is Yeast, throw a soft warning and allows for protocol generation:
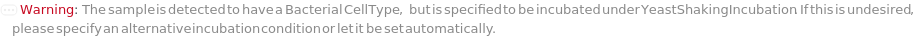
If CellType for samples and SamplesOutStorageCondition are both microbial but one is Bacterial and the other is Yeast, throw a soft warning and allows for protocol generation:
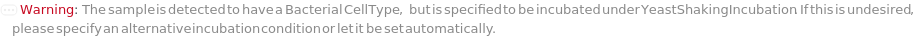
ConflictingCellTypeWithIncubator (2)
If CellType for samples and Incubator are both microbial but one is Bacterial and the other is Yeast, throw a soft warning and allows for protocol generation:

If CellType for samples and Incubator are both microbial but one is Bacterial and the other is Yeast, throw a soft warning and allows for protocol generation:

ConflictingCellTypeWithStorageCondition (3)
If CellType for samples and SamplesOutStorageCondition are both microbial but one is Bacterial and the other is Yeast, throw a soft warning and allows for protocol generation:

If CellType for samples and SamplesOutStorageCondition are both microbial but one is Bacterial and the other is Yeast, throw a soft warning and allows for protocol generation:

If samples are Mammalian, can't specify a Bacterial or Yeast SamplesOutStorageCondition (and vice versa):


ConflictingCultureAdhesion (2)
ConflictingCultureAdhesionWithContainer (2)
A discarded Adherent sample that no longer has a container does not further trigger ConflictingCultureAdhesionWithContainer, or complain about container footprint if thats is the only conflict with the specified Incubator/IncubationCondition, throws DiscardedSample error and returns $Failed:
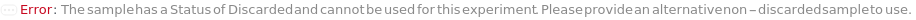




If samples are in a flask, CultureAdhesion can't be SolidMedia:

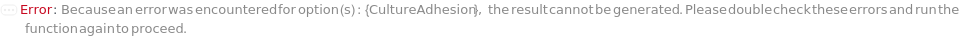
ConflictingIncubationConditionsForSameContainer (1)
ConflictingIncubationConditionWithSettings (2)
ConflictingIncubationWorkCells (1)
ConflictingIncubatorIncubationCondition (1)
ConflictingIncubatorWithSettings (2)
ConflictingShakingConditions (4)
If shaking options are provided, but Shake is set to False, throws an error, returns $Failed, and only uses the provided shaking option values to resolve incubator:


If shaking options are specified to be Null but Shake is set to True, throws an error, returns $Failed, and only uses Shake->True to resolve incubator:
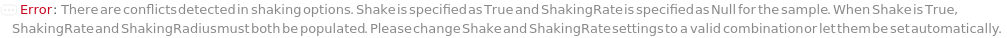

If the two shaking options are specified to be conflicting with each other, throws an error, returns $Failed, and only uses non-null values to resolve incubator:


If using a mammalian sample, and any shaking options are specified to be conflicting, throws an error, returns $Failed, and only uses null values or Shake -> False to resolve incubator:



ConflictingUnitOperationMethodRequirements (7)
If incubator specified is a mix of robotic incubator and manual incubator, throw an error:

If Preparation is set to Robotic but CultureAdhesion is SolidMedia, throw an error:

If Preparation is set to Robotic but IncubationCondition is specified as non-Custom, throw an error:

If Preparation is set to Robotic, then Time must not be greater than 3 hours:

If the CultureAdhesion is detected or specified to be SolidMedia, the preparation is not allowed to be Robotic:

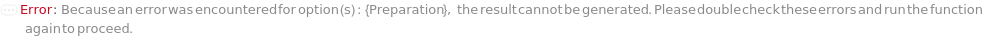
If the QuantificationInstrument is a colony handler while the preparation is Robotic, an error is thrown:


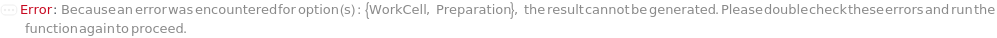
If the QuantificationMethod is ColonyCount while the preparation is Robotic, an error is thrown:

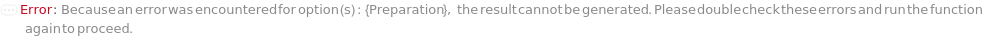
ConflictingWorkCellWithPreparation (2)
If Preparation is set to Manual, WorkCell must not be set. If Preparation is set to Robotic, it must be set to the correct work cell, otherwise an error will be thrown:

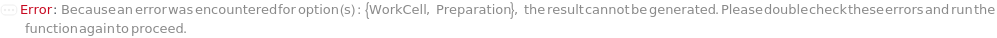
If Preparation is set to Manual, WorkCell must not be set. If Preparation is set to Robotic, it must be set to the correct work cell, otherwise an error will be thrown:

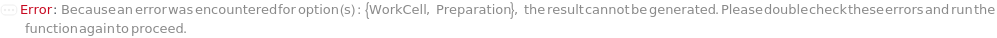
CultureAdhesionNotSpecified (1)
CustomIncubationConditionNotSpecified (1)
EmptySamples (4)
If a sample is liquid but Volume is less than 1% of MaxVolume of container, throws a warning:

If a sample is liquid but Volume is not populated, throws a warning:

If a sample is solid but Mass is less than 1% of MaxVolume of container, throws a warning:
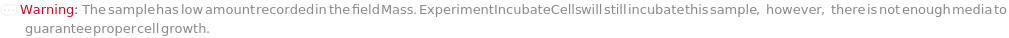
If a sample is solid but Mass is not populated, throws a warning:

IncubationConditionIsIncompatible (4)
If multiple samples have the container and/or cell type not supported by the incubation conditions specified, throws an error and returns $Failed:


If the sample cell type is not supported by the incubation condition specified, throws an error and returns $Failed:


If the sample container is not supported by the incubation condition specified, throws an error and returns $Failed:


If the specified cell type is not supported by the incubation condition specified, throws an error and returns $Failed:



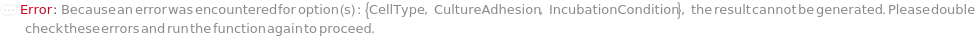
IncubationMaxTemperature (1)
IncubatorIsIncompatible (3)
If a sample's container is incompatible with the incubator model specified, throws an error and returns $Failed:


If CellType is Mammalian, a Bacterial incubator cannot be specified:


If the samples' container, celltype, and/or culture adhesion is not supported by the incubator model specified, throws an error and returns $Failed:


InvalidCellIncubationContainers (2)
InvalidIncubationConditions (1)
InvalidPlateSamples (1)
NoIncubatorForContainersAndSettings (2)
If no incubator can be found to support either the sample (container, cell type, culture adhesion) or the specified incubation settings for multiple samples, throws an error to recommend both transferring the samples and changing options, and returns $Failed:


If no incubator can be found to support either the sample (container, cell type, culture adhesion) or the specified incubation settings, throws an error to recommend both transferring the samples and changing options, and returns $Failed:


NoIncubatorForSettings (2)
If no incubator can be found to support both the sample (container, cell type, culture adhesion) and the specified incubation settings, throws an error to recommend either changing options or alternatively transferring the samples (if applicable) and returns $Failed:

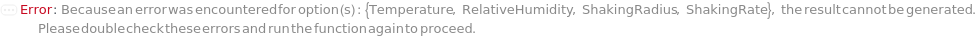
If no incubator can be found to support both the sample (container, cell type, culture adhesion) and the specified incubation settings, throws an error to recommend either changing options or alternatively transferring the samples (if applicable) and returns $Failed:




ObjectDoesNotExist (6)
Do NOT throw a message if we have a simulated container but a simulation is specified that indicates that it is simulated:
Do NOT throw a message if we have a simulated sample but a simulation is specified that indicates that it is simulated:
Throw a message if we have a container that does not exist (ID form):
Throw a message if we have a container that does not exist (name form):
Throw a message if we have a sample that does not exist (ID form):
Throw a message if we have a sample that does not exist (name form):