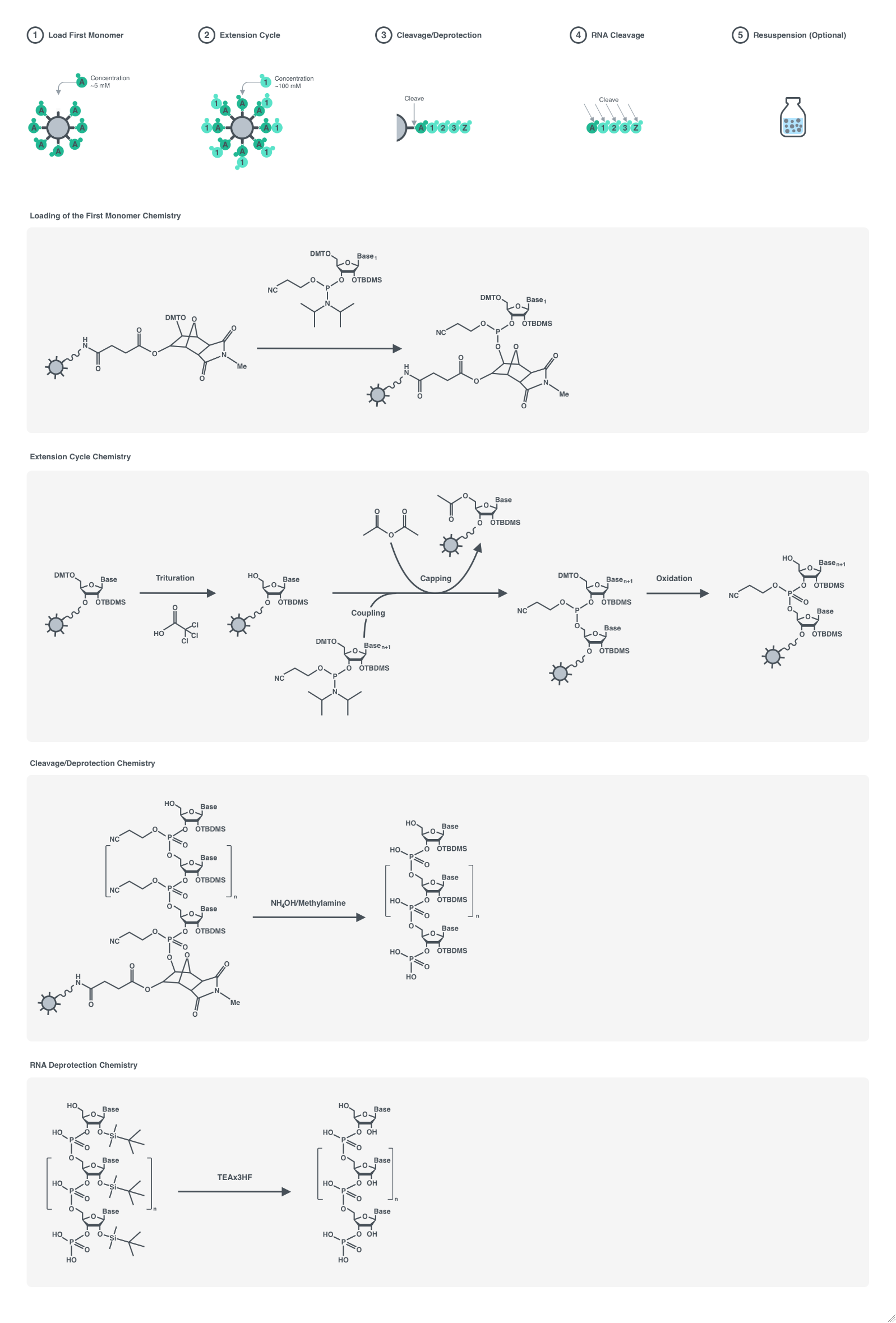
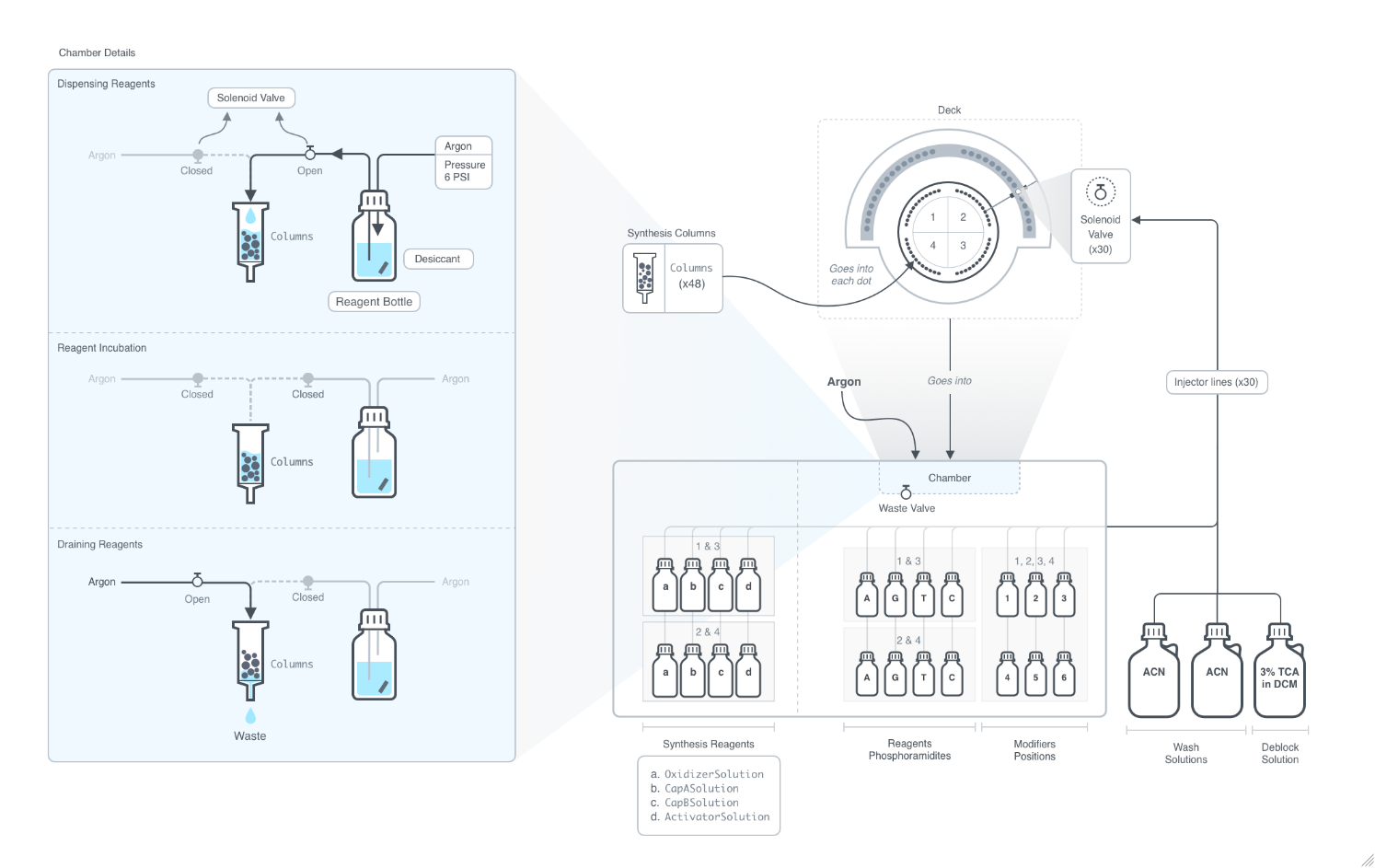
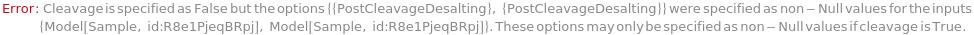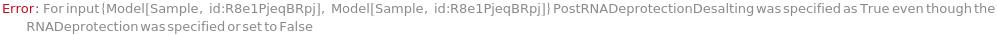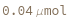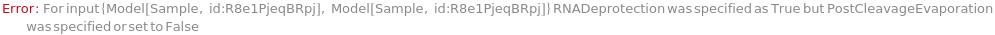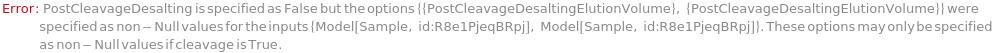Cleavage
PostCleavageDesalting
Indicates if the oligomers are desalted after cleavage.
Default Calculation: Automatically set to False if none of the Desalting options are specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
PostCleavageEvaporation
Indicates if each oligomer is dried after having 2-OH protective group removed.
Default Calculation: Automatically set to True if cleavage is happening.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
Cleavage
Indicates if each oligomer should be cleaved and filtered from the resin after synthesis.
Default Calculation: Automatically set to True unless StorageSolvent or StorageSolventVolume are specified.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
CleavageMethod
The methods defining the cleavage conditions for each oligomer. If a provided method conflicts with the other cleavage options, the specified cleavage options overwrite the corresponding conditions specified by the method.
Default Calculation: If the strand is being cleaved, automatically set to a cleavage method based on the values of the other cleavage options.
Pattern Description: An object of type or subtype Object[Method, Cleavage] or Null.
Programmatic Pattern: ObjectP[{Object[Method, Cleavage]}] | Null
Index Matches to: experiment samples
CleavageTime
The length of time the strands are incubated in cleavage solution.
Default Calculation: If the strand is being cleaved and CleavageMethod is specified, automatically set based on the time used in the specified method. If the strand is being cleaved and CleavageMethod is not specified, automatically set to 8 hours.
Pattern Description: Greater than or equal to 0 hours and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Hour, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
CleavageTemperature
The temperature at which the strands are incubated in cleavage solution.
Default Calculation: If the strand is being cleaved and CleavageMethod is specified, automatically set based on the temperature used in the specified method. If the strand is being cleaved and CleavageMethod is not specified, automatically set to 55C.
Pattern Description: Greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[0*Celsius, 100*Celsius] | Automatic) | Null
Index Matches to: experiment samples
CleavageSolution
The cleavage solution in which the strands cleaved.
Default Calculation: If the strand is being cleaved and CleavageMethod is specified, automatically set based on the cleavage solution used in the specified method. If the strand is being cleaved and CleavageMethod is not specified, automatically set to ammonium hydroxide.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CleavageSolutionVolume
The volume of cleavage solution in which the strands are cleaved.
Default Calculation: If the strand is being cleaved and CleavageMethod is specified, automatically set based on the cleavage solution volume used in the specified method. If the strand is being cleaved and CleavageMethod is not specified, automatically set to 800 uL.
Pattern Description: Greater than or equal to 0 milliliters and less than or equal to 1 milliliter or Null.
Programmatic Pattern: (RangeP[0*Milliliter, 1*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
CleavageWashSolution
The solution used to wash the solid support following cleavage.
Default Calculation: If the strand is being cleaved and CleavageMethod is specified, automatically set based on the cleavage wash solution used in the specified method. If the strand is being cleaved and CleavageMethod is not specified, automatically set to RNAse-free water.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CleavageWashVolume
The volume of solution used to wash the solid support following cleavage.
Default Calculation: If the strand is being cleaved and CleavageMethod is specified, automatically set based on the cleavage solution volume used in the specified method. If the strand is being cleaved and CleavageMethod is not specified, automatically set to 500 uL.
Pattern Description: Greater than or equal to 0 milliliters and less than or equal to 0.8 milliliters or Null.
Programmatic Pattern: (RangeP[0*Milliliter, 0.8*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
StorageSolvent
The solution that uncleaved resin is stored in following synthesis.
Default Calculation: If the strand is not being cleaved, automatically set to water.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
StorageSolventVolume
The amount of solvent that uncleaved resin is stored in post-synthesis.
Default Calculation: If the strand is not being cleaved, automatically set 200 uL for the 40 nmol scale, 400 uL for the 200 nmol scale, 1 mL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 milliliters and less than or equal to 1 milliliter or Null.
Programmatic Pattern: (RangeP[0*Milliliter, 1*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesalting
PostCleavageDesaltingSampleLoadRate
The rate at which each oligomer sample is dispensed into the Cartridge.
Default Calculation: Automatically set to 3 Milliliter/Minute.
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
Nested Index Matches to: experiment samples
PostCleavageDesaltingRinseAndReload
Indicates if each individual sample source well is rinsed with EquilibrationBuffer which is then transferred to the Cartridge that corresponds to the sample pool to improve sample recovery.
Default Calculation: Automatically set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
Nested Index Matches to: experiment samples
PostCleavageDesaltingRinseAndReloadVolume
The volume of PostCleavageDesaltingEquilibrationBuffer added to the source well of each individual oligomer in order to recover more of the sample.
Default Calculation: Automatically set to 0.5*Milliliter if PostCleavageDesaltingRinseAndReload has been specified as True.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 2 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 2*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
Nested Index Matches to: experiment samples
PostCleavageDesaltingType
The type of desalting separation that is used for each oligomer.
Default Calculation: Automatically set to ReversePhase.
Pattern Description: ReversePhase or Null.
Programmatic Pattern: (ReversePhase | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingCartridge
The sorbent-packed container that forms the stationary phase for desalting for each sample pool.
Default Calculation: Automatically set to Model[Container,ExtractionCartridge,"500mg, 3cc, C18, Vac extraction cartridge"].
Pattern Description: An object of type or subtype Object[Container, ExtractionCartridge] or Model[Container, ExtractionCartridge] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Container, ExtractionCartridge], Model[Container, ExtractionCartridge]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingPreFlushVolume
The amount of PostCleavageDesaltingPreFlushBuffer with which each Cartridge is washed prior to oligomer loading. No flush is performed if set to Null.
Default Calculation: If neither the PostCleavageDesaltingPreFlushBuffer nor the corresponding PostCleavageDesaltingPreFlushRate have been specified as Null, the PostCleavageDesaltingPreFlushVolume is automatically set to 5 times of the MaxVolume of the selected Cartridge or 17.5 Milliliter (5 times of the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller. Otherwise, the option is set to Null.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 520 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 520*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingPreFlushRate
The rate at which the PostCleavageDesaltingPreFlushBuffer flows onto the column during the flush step of desalting.
Default Calculation: If neither the PostCleavageDesaltingPreFlushBuffer nor the corresponding PostCleavageDesaltingPreFlushVolume have been specified as Null, the PostCleavageDesaltingPreFlushRate is automatically set to 5 mL/min. Otherwise, the option is set to Null.
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingEquilibrationVolume
The amount of PostCleavageDesaltingEquilibrationBuffer with which each Cartridge is washed prior to oligomer loading.
Default Calculation: The PostCleavageDesaltingEquilibrationVolume is automatically set to 5 times of the MaxVolume of the selected Cartridge or 17.5 Milliliter (5 times of the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 520 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 520*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingEquilibrationRate
The rate at which the PostCleavageDesaltingEquilibrationBuffer flows onto the column during the equilibration step of desalting.
Default Calculation: Automatically set to 3 Milliliter/Minute
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingElutionVolume
The volume of the PostCleavageDesaltingElutionBuffer that is used during the collection step of desalting.
Default Calculation: The PostCleavageDesaltingElutionVolume is automatically set to the MaxVolume of the selected Cartridge or 3.5 Milliliter (the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 3.5 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 3.5*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingElutionRate
The rate at which the PostCleavageDesaltingElutionBuffer flows onto the column during the elution step of desalting.
Default Calculation: Automatically set to 5 Milliliter/Minute
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingWashVolume
The volume of PostCleavageDesaltingWashBuffer that the cartridges are rinsed with after the oligomers are loaded.
Default Calculation: The PostCleavageDesaltingWashVolume is automatically set to 10 times of the MaxVolume of the selected Cartridge or 35 Milliliter (10 times of the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 8000 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 8000*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingWashRate
The rate at which the PostCleavageDesaltingWashBuffer flows onto the column during the equilibration and wash step of desalting.
Default Calculation: Automatically set to 5 Milliliter/Minute.
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostCleavageDesaltingInstrument
The liquid handling instrument used for desalting after cleavage.
Default Calculation: Automatically set to Model[Instrument, LiquidHandler, "GX-271 for Solid Phase Extraction"].
Pattern Description: An object of type or subtype Model[Instrument, LiquidHandler] or Object[Instrument, LiquidHandler]
Programmatic Pattern: ObjectP[{Model[Instrument, LiquidHandler], Object[Instrument, LiquidHandler]}] | Automatic
PostCleavageDesaltingPreFlushBuffer
The solution that is used to wash the sorbent in the Cartridge of any residues before adding the EquilibrationBuffer in desalting steps.
Default Calculation: Automatically set to Model[Sample, StockSolution, "90% methanol"] if PostCleavageDesaltingPreFlushVolume and PostCleavageDesaltingPreFlushRate are not both Null, and to Null otherwise
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
PostCleavageDesaltingEquilibrationBuffer
The solution that is used to wet and condition the sorbent in the Cartridge before oligomers are added to the corresponding Cartridge in order to ensure consistent interaction between the solid phase and the samples.
Default Calculation: Automatically set to Model[Sample, StockSolution, "15 mM ammonium acetate"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
PostCleavageDesaltingWashBuffer
The wash solution that is used in desalting to elute any impurities present in the oligomers that do not stick to the sorbent off the cartridge prior to oligomer elution.
Default Calculation: Automatically set to Model[Sample, "Milli-Q water"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
PostCleavageDesaltingElutionBuffer
The elution solution that is used to disrupt the interaction between the oligomers and the sorbet and elute it into the collection container (Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"]).
Default Calculation: Automatically set to Model[Sample, StockSolution, "90% methanol"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
RNADeprotection
RNADeprotection
Indicates if each oligomer has protection group from 2-OH removed.
Default Calculation: Automatically set to True if strands are being cleaved and no Deprotection options set as Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionMethod
The 2-OH deprotection methods used to specify the 2-OH deprotection conditions for each oligomer. If a provided method conflicts with the other RNA Deprotection options, the specified RNA Deprotection options overwrites the corresponding conditions specified by the method.
Default Calculation: If the strand is having 2-OH protection removed, automatically set to RNA Deprotection method based on the values of the other cleavage options.
Pattern Description: An object of type or subtype Object[Method, RNADeprotection] or Null.
Programmatic Pattern: ObjectP[{Object[Method, RNADeprotection]}] | Null
Index Matches to: experiment samples
RNADeprotectionTime
The length of time the strands are incubated in the RNA Deprotection solution.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the time used in the specified method. If the strand is having 2-OH protection removed and RNADeprotectionMethod is not specified, automatically automatically set to 10 minutes.
Pattern Description: Greater than or equal to 0 hours and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Hour, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionTemperature
The temperature at which the strands are incubated in RNA deprotection solution.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the temperature used in the specified method. If the strand is being cleaved and RNADeprotectionMethod is not specified, automatically set to 65C.
Pattern Description: Greater than or equal to 0 degrees Celsius and less than or equal to 65 degrees Celsius or Null.
Programmatic Pattern: (RangeP[0*Celsius, 65*Celsius] | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionResuspensionSolution
Solvent used to resuspend oligomers after the clevage.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the RNADeprotectionResuspensionSolution used in the specified method. If the strand is having 2-OH protection removed and RNADeprotectionMethod is not specified, automatically set to DMSO.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionResuspensionSolutionVolume
The volume of solvent used to resuspend oligomers after the clevage.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the value from the specified method. If the strand is being cleaved and RNADeprotectionMethod is not specified, automatically set to 100 uL.
Pattern Description: Greater than or equal to 0 milliliters and less than or equal to 1 milliliter or Null.
Programmatic Pattern: (RangeP[0*Milliliter, 1*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionResuspensionTime
The length of time the strands are incubated in the RNA Deprotection resuspension solution.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the time used in the specified method. If the strand is having 2-OH protection removed and RNADeprotectionMethod is not specified, automatically automatically set to 5 minutes.
Pattern Description: Greater than or equal to 0 hours and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Hour, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionResuspensionTemperature
The temperature at which the strands are incubated in RNA deprotection resuspension solution.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the temperature used in the specified method. If the strand is being cleaved and RNADeprotectionMethod is not specified, automatically set to 55C.
Pattern Description: Greater than or equal to 0 degrees Celsius and less than or equal to 65 degrees Celsius or Null.
Programmatic Pattern: (RangeP[0*Celsius, 65*Celsius] | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionSolution
The volume of RNA Deprotection solution used to remove 2-OH protective group.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the RNA Deprotection solution used in the specified method. If the strand is having 2-OH protection removed and RNADeprotectionMethod is not specified, automatically set to TEA with TEAx3HF in DMSO.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionSolutionVolume
The volume of RNA Deprotection solution in which 2-OH protective group is being removed.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the value from the specified method. If the strand is being cleaved and RNADeprotectionMethod is not specified, automatically set to 125 uL.
Pattern Description: Greater than or equal to 0 milliliters and less than or equal to 1 milliliter or Null.
Programmatic Pattern: (RangeP[0*Milliliter, 1*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionQuenchingSolution
The solution for quenching the 2-OH protective group deprotection reaction.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the RNADeprotectionQuenchingSolution used in the specified method. If the strand is having 2-OH protection removed and RNADeprotectionMethod is not specified, automatically set to TRIS buffer.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
RNADeprotectionQuenchingSolutionVolume
The volume of RNA Deprotection Quenching solution used to stop the deprotection reaction.
Default Calculation: If the strand is having 2-OH protection removed and RNADeprotectionMethod is specified, automatically set based on the value from the specified method. If the strand is being cleaved and RNADeprotectionMethod is not specified, automatically set to 1750 uL.
Pattern Description: Greater than or equal to 0 milliliters and less than or equal to 1.75 milliliters or Null.
Programmatic Pattern: (RangeP[0*Milliliter, 1.75*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionEvaporation
Indicates if each oligomer is dried after having 2-OH protective group removed.
Default Calculation: Automatically set to True if RNADeprotection is happening.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesalting
Indicates if the oligomers should be desalted after removing 2-OH protective group and drying.
Default Calculation: Automatically set to False unless related options are specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesalting
PostRNADeprotectionDesaltingSampleLoadRate
The rate at which each oligomer sample is dispensed into the Cartridge.
Default Calculation: Automatically set to 3 Milliliter/Minute.
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
Nested Index Matches to: experiment samples
PostRNADeprotectionDesaltingRinseAndReload
Indicates if each individual sample source well is rinsed with EquilibrationBuffer which is then transferred to the Cartridge that corresponds to the sample pool to improve sample recovery.
Default Calculation: Automatically set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
Nested Index Matches to: experiment samples
PostRNADeprotectionDesaltingRinseAndReloadVolume
The volume of PostRNADeprotectionDesaltingEquilibrationBuffer added to the source well of each individual oligomer in order to recover more of the sample.
Default Calculation: Automatically set to 0.5*Milliliter if PostRNADeprotectionDesaltingRinseAndReload has been specified as True.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 2 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 2*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
Nested Index Matches to: experiment samples
PostRNADeprotectionDesaltingType
The type of desalting separation that is used for each oligomer.
Default Calculation: Automatically set to ReversePhase.
Pattern Description: ReversePhase or Null.
Programmatic Pattern: (ReversePhase | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingCartridge
The sorbent-packed container that forms the stationary phase for desalting for each sample pool.
Default Calculation: Automatically set to Model[Container,ExtractionCartridge,"500mg, 3cc, C18, Vac extraction cartridge"].
Pattern Description: An object of type or subtype Object[Container, ExtractionCartridge] or Model[Container, ExtractionCartridge] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Container, ExtractionCartridge], Model[Container, ExtractionCartridge]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingPreFlushVolume
The amount of PostRNADeprotectionDesaltingPreFlushBuffer with which each Cartridge is washed prior to oligomer loading. No flush is performed if set to Null.
Default Calculation: If neither the PostRNADeprotectionDesaltingPreFlushBuffer nor the corresponding PostRNADeprotectionDesaltingPreFlushRate have been specified as Null, the PostRNADeprotectionDesaltingPreFlushVolume is automatically set to 5 times of the MaxVolume of the selected Cartridge or 17.5 Milliliter (5 times of the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller. Otherwise, the option is set to Null.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 520 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 520*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingPreFlushRate
The rate at which the PostRNADeprotectionDesaltingPreFlushBuffer flows onto the column during the flush step of desalting.
Default Calculation: If neither the PostRNADeprotectionDesaltingPreFlushBuffer nor the corresponding PostRNADeprotectionDesaltingPreFlushVolume have been specified as Null, the PostRNADeprotectionDesaltingPreFlushRate is automatically set to 5 mL/min. Otherwise, the option is set to Null.
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingEquilibrationVolume
The amount of PostRNADeprotectionDesaltingEquilibrationBuffer with which each Cartridge is washed prior to oligomer loading.
Default Calculation: The PostRNADeprotectionDesaltingEquilibrationVolume is automatically set to 5 times of the MaxVolume of the selected Cartridge or 17.5 Milliliter (5 times of the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 520 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 520*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingEquilibrationRate
The rate at which the PostRNADeprotectionDesaltingEquilibrationBuffer flows onto the column during the equilibration step of desalting.
Default Calculation: Automatically set to 3 Milliliter/Minute
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingElutionVolume
The volume of the PostRNADeprotectionDesaltingElutionBuffer used during the collection step of desalting.
Default Calculation: The PostRNADeprotectionDesaltingElutionVolume is automatically set to the MaxVolume of the selected Cartridge or 3.5 Milliliter (the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 3.5 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 3.5*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingElutionRate
The rate at which the PostRNADeprotectionDesaltingElutionBuffer flows onto the column during the elution step of desalting.
Default Calculation: Automatically set to 5 Milliliter/Minute
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingWashVolume
The volume of PostRNADeprotectionDesaltingWashBuffer that the cartridges is rinsed with after the oligomers are loaded.
Default Calculation: The PostRNADeprotectionDesaltingWashVolume is automatically set to 10 times of the MaxVolume of the selected Cartridge or 35 Milliliter (10 times of the MaxVolume of Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"], the collection plate for eluted samples from SPE cartridges), whichever is smaller.
Pattern Description: Greater than or equal to 0.1 milliliters and less than or equal to 8000 milliliters or Null.
Programmatic Pattern: (RangeP[0.1*Milliliter, 8000*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingWashRate
The rate at which the PostRNADeprotectionDesaltingWashBuffer flows onto the column during the equilibration and wash step of desalting.
Default Calculation: Automatically set to 5 Milliliter/Minute.
Pattern Description: Greater than or equal to 0.01 milliliters per minute and less than or equal to 45 milliliters per minute or Null.
Programmatic Pattern: (RangeP[0.01*(Milliliter/Minute), 45*(Milliliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
PostRNADeprotectionDesaltingInstrument
The liquid handling instrument used for desalting after removing protection from 2-OH group.
Default Calculation: Automatically set to Model[Instrument, LiquidHandler, "GX-271 for Solid Phase Extraction"].
Pattern Description: An object of type or subtype Model[Instrument, LiquidHandler] or Object[Instrument, LiquidHandler]
Programmatic Pattern: ObjectP[{Model[Instrument, LiquidHandler], Object[Instrument, LiquidHandler]}] | Automatic
PostRNADeprotectionDesaltingPreFlushBuffer
The solution that is used to wash the sorbent in the Cartridge of any residues before adding the EquilibrationBuffer in desalting steps.
Default Calculation: Automatically set to Model[Sample, StockSolution, "90% methanol"] if PostRNADeprotectionDesaltingPreFlushVolume and PostRNADeprotectionDesaltingPreFlushRate are not both Null, and to Null otherwise
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
PostRNADeprotectionDesaltingEquilibrationBuffer
The solution that is used to wet and condition the sorbent in the Cartridge before oligomers are added to the corresponding Cartridge in order to ensure consistent interaction between the solid phase and the samples.
Default Calculation: Automatically set to Model[Sample, StockSolution, "15 mM ammonium acetate"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
PostRNADeprotectionDesaltingWashBuffer
The wash solution that is used in desalting to elute any impurities present in the oligomers that do not stick to the sorbent off the cartridge prior to oligomer elution.
Default Calculation: Automatically set to Model[Sample, "Milli-Q water"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
PostRNADeprotectionDesaltingElutionBuffer
The elution solution that is used to disrupt the interaction between the oligomers and the sorbet and elute it into the collection container (Model[Container, Plate, "48-well Pyramid Bottom Deep Well Plate"]).
Default Calculation: Automatically set to Model[Sample, StockSolution, "90% methanol"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
Protocol
Instrument
The instrument used to perform the synthesis.
Default Value: Model[Instrument, DNASynthesizer, ABI 3900]
Pattern Description: An object of type or subtype Model[Instrument, DNASynthesizer] or Object[Instrument, DNASynthesizer]
Programmatic Pattern: ObjectP[{Model[Instrument, DNASynthesizer], Object[Instrument, DNASynthesizer]}]
Scale
The theoretical amount of reaction sites available for synthesis.
Default Calculation: If Column is specified as an object, automatically set to the amount of reaction sites on the resin. Otherwise, automatically set to 0.2 Micromole
Pattern Description: 40 nanomoles, 0.2 micromoles, or 1 micromole.
Programmatic Pattern: (40*Nanomole | 0.2*Micromole | 1*Micromole) | Automatic
Columns
The solid support on which the synthesis is carried out.
Default Value: Model[Sample, UnySupport]
Pattern Description: An object of type or subtype Model[Sample], Object[Sample], or Model[Resin] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample], Model[Resin]}] | _String
Index Matches to: experiment samples
NumberOfReplicates
Number of times each of the input strands should be synthesized using identical experimental parameters.
Pattern Description: Greater than or equal to 1 in increments of 1 or Null.
Programmatic Pattern: GreaterEqualP[1, 1] | Null
Phosphoramidites
Phosphoramidites
The phosphoramidite solutions used for each monomer.
Default Calculation: Set to default Stock Solution specified in SyntheticMonomers field of Model[Physics, Oligomer] for this type of oligomer.
Pattern Description: List of one or more {Monomer, Phosphoramidite} entries.
Programmatic Pattern: {{SequenceP, ObjectP[Model[Sample, StockSolution]] | {{ObjectP[Model[Sample, StockSolution]], RangeP[0.01, 1.]}..} | {{SequenceP, RangeP[0.01, 1.]}..}}..} | Automatic
PhosphoramiditeDesiccants
Indicates if desiccant packets should be added to the phosphoramidite bottles.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
Initial Wash
NumberOfInitialWashes
The number of washes at the start of the synthesis.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1]
InitialWashTime
The wait time between the washes at the start of the synthesis.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
InitialWashVolume
The volume of each wash at the start of the synthesis.
Default Calculation: Automatically set to 200 uL for the 40 nmol scale, 250 uL for the 200 nmol scale, 280 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
Deprotection
NumberOfDeprotections
The number of times that the detritylation solution is added to the resin after each cycle.
Default Calculation: Automatically set to 2 for 40 nmol and 200 nmol scales; automatically set to 3 for 1 umol scale.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1] | Automatic
DeprotectionTime
The wait time between each detritylation iteration.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
DeprotectionVolume
The volume of detritylation solvent used in each detritylation iteration.
Default Calculation: automatically set to 60 uL for the 40 nmol scale, 140 uL for the 200 nmol scale, 180 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
NumberOfDeprotectionWashes
The number of times the resin is washed after detritylation.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1]
DeprotectionWashTime
The wait time between the post-detritylation washes.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
DeprotectionWashVolume
The volume of each post-detritylation wash.
Default Calculation: Automatically set to 200 uL for the 40 nmol scale, 250 uL for the 200 nmol scale, 280 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
FinalDeprotection
Indicates if a final deprotection (detritylation) step is done following the last synthesis cycle.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
Index Matches to: experiment samples
Coupling
ActivatorSolution
The solution that interacts with the the phosphoramidite prior to attaching next monomer by protonating the diisopropylamino group of the nucleoside phosphoramidite, increasing the coupling reaction efficiency, typically a tetrazole derivative dissolved in acetonitrile.
Default Calculation: Automatically set Model[Sample, "Activator"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Automatic or list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Automatic entries.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | {((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic)..} | Automatic) | Automatic
ActivatorVolume
The volume of activator solution used to increase coupling reaction efficiency of the amidite prior to attaching next monomer.
Default Calculation: automatically set to 40 uL for the 40 nmol scale, 45 uL for the 200 nmol scale, 115 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
PhosphoramiditeVolume
The volume of phosphoramidite solution used in each attaching next monomer iteration for each monomer. A,U,G,C (specified by "Natural") must use the same phosphoramidite volume.
Default Calculation: Automatically set to 20 uL for the 40 nmol scale, 30 uL for the 200 nmol scale, 75 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 280 microliters or list of one or more {Monomer, Volume} entries.
Programmatic Pattern: (RangeP[1*Microliter, 280*Microliter] | {{Modification[_] | "Natural", RangeP[0*Microliter, 280*Microliter]}..}) | Automatic
NumberOfCouplings
The number of next monomer attachment iterations for each monomer type. A,U,G,C (specified by "Natural") must use the same number of couplings.
Default Calculation: Automatically set to 1 for the 40 nmol scale, 2 for the 200 nmol scale, 3 for the 1 umol scale.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1 or list of one or more {Monomer, Couplings} entries.
Programmatic Pattern: (RangeP[1, 5, 1] | {{Modification[_] | "Natural", RangeP[1, 5, 1]}..}) | Automatic
CouplingTime
The wait time between each next monomer attachment iteration for each monomer. A,U,G,C (specified by "Natural") must use the same next monomer attachment time.
Default Calculation: Automatically set to 6 minutes.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 30 minutes or list of one or more {Monomer, Time} entries.
Programmatic Pattern: (RangeP[0*Second, 30*Minute] | {{Modification[_] | "Natural", RangeP[0*Second, $MaxExperimentTime]}..}) | Automatic
Capping
CapASolution
The solution (used in conjunction with Cap B Solution) used to passivate unreacted sites by acetylating free hydroxy sites, typically a mixture of tetrahydrofuran, lutidine, and acid anhydride.
Default Calculation: For RNA defaults to Ultramild Cap A, otherwise, defaults Cap A (based on acetic anhydride).
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Automatic or list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Automatic entries.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | {((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic)..} | Automatic) | Automatic
CapBSolution
The solution (used in conjunction with Cap A Solution) used to passivate unreacted sites by acetylating free hydroxy sites, typically a solution of 1-methylimidazole in tetrahydrofuran.
Default Calculation: Automatically default to Model[Sample, "Capping solution B"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Automatic or list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Automatic entries.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | {((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic)..} | Automatic) | Automatic
NumberOfCappings
The number of passivation iterations on each synthesis cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1]
CapTime
The wait time between each passivation iteration.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
CapAVolume
The volume of Cap A solution used in each passivation iteration.
Default Calculation: Automatically set to 20 uL for the 40 nmol scale, 30 uL for the 200 nmol scale, 80 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
CapBVolume
The volume of Cap B solution used in each passivation iteration.
Default Calculation: Automatically set to 20 uL for the 40 nmol scale, 30 uL for the 200 nmol scale, 80 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
Oxidation
OxidationSolution
The solution used to oxidize phosphorus(III) linker into phosphate, typically a solution of Iodine in THF with Pyridine and water.
Default Value: Model[Sample, Oxidizer]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
NumberOfOxidations
The number of oxidation iterations.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1]
OxidationTime
The wait time between each oxidation iteration.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
OxidationVolume
The volume of oxidation solution used in each oxidation iteration.
Default Calculation: Automatically set to 30 uL for the 40 nmol scale, 60 uL for the 200 nmol scale, 150 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
NumberOfOxidationWashes
The number of washes following oxidation.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1]
OxidationWashTime
The wait time between each post-oxidation wash.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
OxidationWashVolume
The volume of wash solution for each post-oxidation wash.
Default Calculation: Automatically set to 200 uL for the 40 nmol scale, 250 uL for the 200 nmol scale, 280 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
SecondaryOxidationSolution
The solution used to oxidize phosphorus(III) linker after coupling in the secondary set of cycle parameters.
Default Calculation: If SecondaryCyclePositions is specified as Null, resolves to Null, otherwise resolves to Model[Sample, "Oxidizer"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
SecondaryNumberOfOxidations
The number of oxidation iterations in the secondary set of cycle parameters.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1]
SecondaryOxidationTime
The wait time between each oxidation iteration in the secondary set of cycle parameters.
Default Value: 60 seconds
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 20 minutes.
Programmatic Pattern: RangeP[0*Second, 20*Minute]
SecondaryOxidationVolume
The volume of oxidation solution used in each oxidation iteration in the secondary set of cycle parameters.
Default Calculation: Automatically set to 30 uL for the 40 nmol scale, 60 uL for the 200 nmol scale, 150 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
SecondaryNumberOfOxidationWashes
The number of washes following oxidation in the secondary set of cycle parameters.
Pattern Description: Greater than or equal to 1 and less than or equal to 5 in increments of 1.
Programmatic Pattern: RangeP[1, 5, 1]
SecondaryOxidationWashTime
The wait time between each post-oxidation wash in the secondary set of cycle parameters.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
SecondaryOxidationWashVolume
The volume of wash solution for each post-oxidation wash in the secondary set of cycle parameters.
Default Calculation: Automatically set to 200 uL for the 40 nmol scale, 250 uL for the 200 nmol scale, 280 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
SecondaryCyclePositions
Positions that should use the secondary cycle parameters (Lower Case).
Pattern Description: An expression that matches the pattern: ListableP[_Integer] or Null.
Programmatic Pattern: ListableP[_Integer] | Null
Index Matches to: experiment samples
Final Wash
NumberOfFinalWashes
The number of washes at the end of the synthesis.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1.
Programmatic Pattern: RangeP[1, 10, 1]
FinalWashTime
The wait time between the washes at the end of the synthesis.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime]
FinalWashVolume
The volume of each wash at the start of the synthesis.
Default Calculation: Automatically set to 200 uL for the 40 nmol scale, 250 uL for the 200 nmol scale, 280 uL for the 1 umol scale.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 280 microliters.
Programmatic Pattern: RangeP[0*Microliter, 280*Microliter] | Automatic
Post Experiment
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples