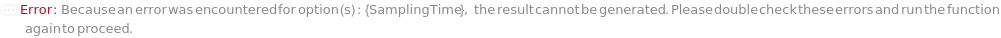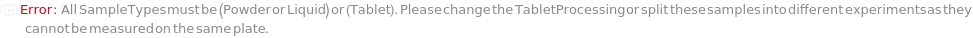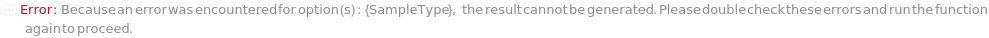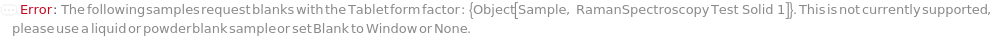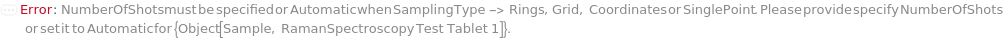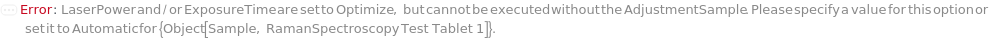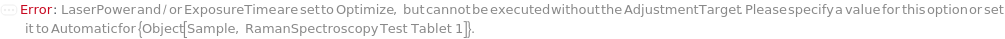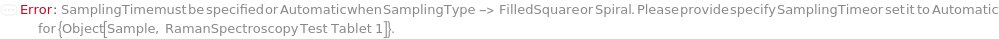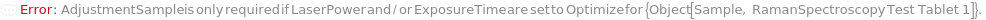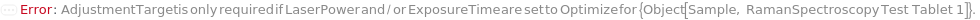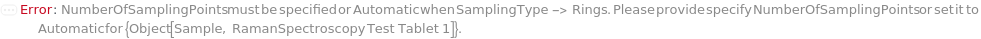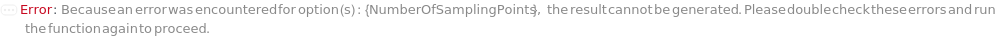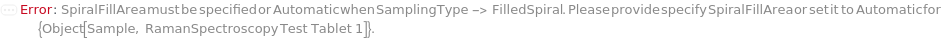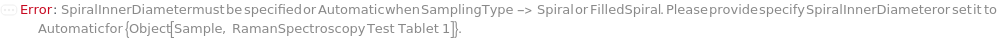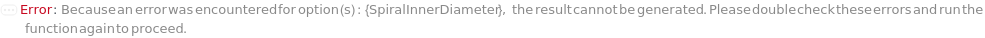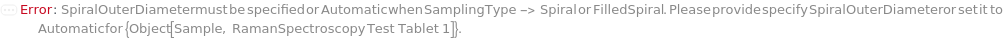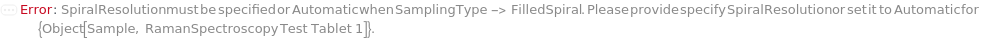ExperimentRamanSpectroscopy
ExperimentRamanSpectroscopy[Samples]⟹Protocol
generates a Protocol object for performing Raman Spectroscopy on the provided Samples in the THz and IR region (10 cm-1 to 3800 cm-1).
Raman spectroscopy is a generally non-destructive analytical technique which provides information about chemical structure and bulk characteristics including morphology, phase, crystallinity, and intermolecular interactions. It is based on the scattering of intense single wavelength light, which is dependent on the interaction of the incident light with chemical bonds within the sample. Stokes and anti-Stokes scattering give information about the spacing of the energetic states in the sample, while the Rayleigh scattering is removed to allow sensitivity in the THz region. Transitions with energies in the THZ or IR region (10 cm-1 to 3800 cm -1) are recorded as peaks on the Raman spectrum with their frequencies reported relative to the excitation frequency. Only transitions which result in change in polarizablity will be visible with this technique; Raman-silent modes can generally be seen by IR spectroscopy. This technique can be applied to crude or purified solids and liquids, and provides information about sample identity, sample purity, as well as sample homogeneity. The experimental results include an averaged spectrum for the entire sample and spatially resolved Raman response generated by measurements at specified coordinates or along a sampling pattern.
Experimental Principles
Figure 1.1: Procedural overview of a Raman Spectroscopy experiment. Step 1: Solid, liquid, or tablet samples are prepared and moved into the sample plate, which is then placed on the sample stage. Step 2: Appropriate adjustments are made to the input power, focus and exposure to maximize the signal-to-noise ratio without saturating the detector. The sample is then irradiated with visible or near-IR light to induce Raman scattering. Step 3: The Raman spectrum is measured over the low frequency/THz region and the IR region, giving information about the structural and chemical composition of the sample respectively. Measurement is performed at a single point or as a function of location by scanning over individual wells or multiple wells in the XY direction. Spatially resolved Raman signals from mapping measurements can be used for principle component analysis based on specific peak intensities.
Instrumentation
THz Raman WPS
Figure 2.1: The THz-Raman Well Plate System (WPS) is a plate-based Raman spectrometer composed of a laser source, focusing and filtering optics, a sample stage, and detector. Panel 1: The input signal is generated with a continuous wave diode laser (785 nm, up to 400 mW). Input light is filtered to remove stray frequency photons and directed towards the bottom of the sample plate via a beamsplitter. The input is focused on the bottom of the sample plate using a 2x, 4x, 10x, or 20x objective. Backscattered light from the sample is collected and passed through a pinhole aperture to remove out of focus signal, then filtered through a narrow bandpass notch filter to reduce the intensity of the Rayleigh scattering. Panel 2: The filtered signal is transmitted to a spectrometer module, composed of gratings and a CCD detector, which records the intensity of scattered light from the THz (5 wavenumber) to IR (2800 wavenumber) regime. The Raman spectrum is plotted as scattering intensity against frequency of the scattered light referenced to the Rayleigh frequency. Scattering with higher energy than the Rayleigh line (anti-Stokes scattering) is recorded on the negative axis to -800 wavenumber, while lower energy scattering (Stokes scattering) is recorded on the positive axis to 2800 wavenumber. Raman maps are generated using the relative intensity of constituent scattering peaks measured at each spatial position. Panel 3: The sample stage can be translated in the X, Y, and Z axes, allowing the instrument to scan using predetermined patterns within an individual well or between multiple wells. An optical camera is also mounted under the motorized sample stage to image the sample and identify areas from which data was collected.
Experiment Options
General
SampleType
For each member of SamplesIn, specifies the form factor of the sample as liquid or solid (powder or tablet) which will occupy a well in the sample plate.
Default Calculation: The sample type is determined from the State and Tablet fields of the Object[Sample], and by Aliquot options specifying dilution and the TabletProcessing option. For example, a solid sample with TargetConcentration specified will have SampleType -> Liquid, and a tablet sample with TabletProcessing -> Grind will have SampletType -> Solid.
Instrument
Pattern Description: An object of type or subtype Model[Instrument, RamanSpectrometer] or Object[Instrument, RamanSpectrometer]
Programmatic Pattern: ObjectP[{Model[Instrument, RamanSpectrometer], Object[Instrument, RamanSpectrometer]}]
TabletProcessing
For each member of SamplesIn, specifies if a tablet should be ground or cut prior to analysis. LargestCrossSection and SmallestCrossSection specify that the tablet is to be cut in half along an axis to expose either the largest or smallest amount of the tablet interior. The largest cross section is obtained by cleaving the tablet parallel to the longest axis, while the smallest cross section will be obtained by cleaving the tablet perpendicular to the longest axis. For scored round tablets, the direction of the score is considered the long axis, and for unscored round tablets LargestCrossSection and SmallestCrossSection are equivalent. The cut face of the tablet is placed facing downward and is subjected to Raman analysis.
Default Calculation: If SampleType -> Solid for a Tablet input, TabletProcessing is set to Grind. For Tablet samples with SampleType -> Automatic or Tablet, TabletProcessing is set to Whole and for all other samples it is set to Null.
Programmatic Pattern: ((Whole | LargestCrossSection | SmallestCrossSection | Grind) | Automatic) | Null
Data Processing
Blank
For each member of SamplesIn, the source used to generate a blank sample whose Raman scattering response is subtracted as background from every Raman spectrum collected for that sample. Window indicates that a stored spectrum of the window material wil be subtracted, while None indicates that there will be no blank subtraction.
Default Calculation: Automatically set to Window for samples with SampleType -> Liquid or Powder, and None when SampleType -> Tablet.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | (Window | None)) | Automatic
Optics
FloodLight
For each member of SamplesIn, indicates if spectroscopic measurements are performed with an objective that creates a broad spot size of approximately 1 Millimeter. FloodLight is best used to obtain a rapid average over the entire sample in cases where spatial resolution of the Raman scattering is not the primary goal.
ObjectiveMagnification
For each member of SamplesIn, specifies the magnification factor objective to be used for the excitation beam, optical imaging, and Raman backscattering.
Default Calculation: If FloodLight is True, the ObjectiveMagnification will be set to Null if no optical image is requested, and 2X if optical images are to be taken along with spectroscopic measurements. Otherwise ObjectiveMagnification is set to 10.
LaserPower
For each member of SamplesIn, specifies the percent of the maximum laser output (400 mW) at the objective. Insufficient LaserPower may result in low signal-to-noise ratio, while excessively LaserPower can result in detector saturation (and loss or spectral resolution) and sample damage. If you are unsure of an appropriate LaserPower, select Optimize.
Pattern Description: Greater than or equal to 1 percent and less than or equal to 100 percent or Optimize.
ExposureTime
For each member of SamplesIn, specifies the amount of time over which the Raman scattering signal is summed by the CCD detector for each measurement. Insufficient ExposureTime results in weak signal, while excessive ExposureTime may lead to detector saturation and loss of spectral resolution. If you are unsure of an appropriate ExposureTime, select Optimize.
AdjustmentSample
For each member of SamplesIn, specifies the sample that should be used to adjust the LaserPower and ExposureTime. If All is selected, the LaserPower and ExposureTime will be adjusted for each sample individually.
Default Calculation: If LaserPower or ExposureTime are set to Optimize, the first measured sample will be used as the adjustment sample.
Programmatic Pattern: ((All | (ObjectP[{Object[Sample], Model[Sample]}] | _String)) | Automatic) | Null
AdjustmentEmissionWavelength
For each member of SamplesIn, specifies the Raman shift wavelength to be used for optimizing signal intensity. "Max" indicates that the most intense scattering peak will be used.
Default Calculation: If AdjustmentSample is specified or All, AdjustmentEmissionWavelength will be set to use the most intense scattering peak. Otherwise, this option will be set to Null.
Programmatic Pattern: ((Max | (RangeP[-600*(1/Centimeter), -10*(1/Centimeter)] | RangeP[10*(1/Centimeter), 3800*(1/Centimeter)])) | Automatic) | Null
AdjustmentTarget
For each member of SamplesIn, specifies the target intensity for an optimized signal as a percentage of the saturation threshold of the detector.
Default Calculation: If AdjustmentSample is specified or All, or Optimize is indicated in ExposureTime or LaserPower, AdjustmentTarget is set to 50 Percent, otherwise this option is set to Null.
Pattern Description: Greater than or equal to 1 percent and less than or equal to 100 percent or Null.
BackgroundRemove
For each member of SamplesIn, indicates if a dark background is subtracted from the sample spectrum. The background is collected with the beam shutter closed and accounts for noise in the CCD detector.
CosmicRadiationFilter
For each member of SamplesIn, indicates if spectra are processed to remove features arising from cosmic rays. Cosmic rays are random and intense bursts of radiation that overwhelm the Raman scattering signal. It is highly recommended that this option is used to eradicate cosmic radiation features which can obfuscate the sample scatting. Note that NumberOfShots should be set to more than 1 (if applicable) in order for this feature to function optimally.
CalibrationCheck
Indicates if the instrument collects a spectrum using a PMMA internal standard prior to measuring the samples. The calibration check reports a value from 0 to 1 representing the similarity between the collected spectrum and the manufacturer collected spectrum (1 is a perfect match). Note that this assessment will not adjust the instrument in any way.
Optical Imaging
InvertedMicroscopeImage
For each member of SamplesIn, indicates if an optical image of the sample is taken using an inverted microscope configuration. The optical camera will utilize ObjectiveMagnification.
Default Calculation: If MicroscopeImageExposureTime or MicroscopeImageLightIntensity are informed, InvertedMicroscopeImage is set to True.
MicroscopeImageExposureTime
For each member of SamplesIn, specifies the exposure time for optical microscope images. Optimize will automatically adjust the exposure for best image quality.
MicroscopeImageLightIntensity
For each member of SamplesIn, specifies the power of the blue LED light that illuminates the sample stage for optical imaging.
Reading
ReadPattern
NumberOfReads
For each member of SamplesIn, specifies the number of times that the SamplingPattern is repeated on the sample.
ReadRestTime
For each member of SamplesIn, specifies the delay between executing the SamplingPattern in a given well.
Sampling
SamplingPattern
For each member of SamplesIn, specifies pattern or set of points that are samples within each well. SinglePoint: Collects spectra at a single point of the well. Spiral: Collects spectra at each point along a spiral pattern. FilledSpiral: Collects spectra at each point along a spiral pattern with the specified area coverage. FilledSquare: Collects spectra in a designated square area by sampling along an S-pattern. Grid: Collects spectra along specified grid lines. Rings: Collects spectra at sparse points on nested rings. Custom: Collects spectra at the requested coordinates.
Figure 3.1: Specific motion patterns can be indicated using the available SamplingPattern set and associated options for each pattern. By mapping over the sample, it is possible to compose images of the sample based on the Raman spectra collected at each point. Patterns are restricted to the well in which the sample is located, though it is possible to specify the z-dimension in the Grid and Coordinates patterns to analyze the sample at a variety of depths.
Programmatic Pattern: SinglePoint | Spiral | FilledSpiral | FilledSquare | Grid | Rings | Coordinates
SamplingTime
For each member of SamplesIn, specifies the amount of time to complete the requested Spiral or Grid SamplingPattern.
Default Calculation: The value of SamplingTime is based on the SamplingPattern. For SamplingPattern -> (Spiral or FilledSquare) this will be set to 600 Second.
Pattern Description: Greater than or equal to 1 second and less than or equal to 9999 seconds or Null.
SpiralInnerDiameter
For each member of SamplesIn, specifies the inner diameter of the Spiral or FilledSpiral SamplingPattern.
Default Calculation: If the SamplingPattern is set Spiral or Filled Spiral, SpiralInnerDiameter will be set to 50 Micrometer if SpiralOuterDiameter is Automatic or greater than 100 Micrometer, and 1 Micrometer is SpiralOuterDiameter is less than 100 Micrometer.
Pattern Description: Greater than or equal to 10 micrometers and less than or equal to 250000 micrometers or Null.
SpiralOuterDiameter
For each member of SamplesIn, specifies the outer diameter of the Spiral or FilledSpiral SamplingPattern.
Default Calculation: If the SamplingPattern is set Spiral or FilledSpiral, SpiralOuterDiameter will be set to 200 Micrometer if SpiralInnerDiameter is Automatic or less than 100 Micrometer, and SpiralInnerDiameter + 200 Micrometer otherwise.
Pattern Description: Greater than or equal to 10 micrometers and less than or equal to 250000 micrometers or Null.
SpiralFillArea
For each member of SamplesIn, specifies the percentage of the total area inside the FilledSpiral pattern from which spectra are collected. Note that when percentages greater than 100 Percent are specified, the pattern will include overlapping measurements.
Default Calculation: If the SamplingPattern is set to FilledSpiral, SpiralFillArea will be set to 50 Percent.
Pattern Description: Greater than or equal to 1 percent and less than or equal to 999 percent or Null.
SpiralResolution
For each member of SamplesIn, specifies the distance between adjacent sampling points in the FilledSpiral or Spiral pattern.
Default Calculation: If the SamplingPattern is set to Spiral or FilledSpiral, SpiralResolution will be set to 10 Micrometer.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 250000 micrometers or Null.
SamplingXDimension
For each member of SamplesIn, specifies the size of the Grid or FilledSquare SamplingPattern in the X direction.
Default Calculation: If the SamplingPattern is set to Grid or FilledSquare, SamplingXDimension will be set to 200 Micrometer.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 250000 micrometers or Null.
SamplingYDimension
For each member of SamplesIn, specifies the size of the Grid or FilledSquare SamplingPattern in the Y direction.
Default Calculation: If the SamplingPattern is set to Grid or FilledSquare, SamplingYDimension will be set to 200 Micrometer.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 250000 micrometers or Null.
SamplingZDimension
Default Calculation: When SamplingPattern -> Grid, this will be set to 0 Micrometer, indicating that no z steps will be taken.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 2500 micrometers or Null.
SamplingXStepSize
For each member of SamplesIn, specifies the spacing between adjacent points of the Grid SamplingPattern in the X direction.
Default Calculation: If the SamplingPattern is set to Grid, SamplingXStepSize will be set to 20 Micrometer.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 250000 micrometers or Null.
SamplingYStepSize
For each member of SamplesIn, specifies the spacing between adjacent points of the Grid SamplingPattern in the Y direction.
Default Calculation: If the SamplingPattern is set to Grid, SamplingYStepSize will be set to 20 Micrometer.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 250000 micrometers or Null.
SamplingZStepSize
For each member of SamplesIn, specifies the spacing between adjacent points of the Grid SamplingPattern in the Z direction.
Default Calculation: When SamplingPattern -> Grid, this will be set to 0 Micrometer, indicating that no step will be taken.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 2500 micrometers or Null.
FilledSquareNumberOfTurns
For each member of SamplesIn, specifies the number of parallel lines within the FilledSquare pattern along which spectra are recorded.
Default Calculation: If the SamplingPattern is set to FilledSquare, FilledSquareNumberOfTurns is set to 10.
Pattern Description: Greater than or equal to 1 and less than or equal to 999 in increments of 1 or Null.
RingSpacing
For each member of SamplesIn, specifies the difference in radius of consecutive nested rings for the Rings SamplingPattern. This value also designates the radius of the innermost ring.
Default Calculation: If the SamplingPattern is set to Rings, RingSpacing will be set to 100 Micrometer.
Pattern Description: Greater than or equal to 1 micrometer and less than or equal to 250000 micrometers or Null.
NumberOfRings
For each member of SamplesIn, specifies the number of evenly spaced rings in the Rings SamplingPattern.
Pattern Description: Greater than or equal to 1 and less than or equal to 99 in increments of 1 or Null.
NumberOfSamplingPoints
For each member of SamplesIn, specifies the total number of distinct measurement points generated along the circumference of the rings specified by NumberOfRings and RingSpacing.
Pattern Description: Greater than or equal to 1 and less than or equal to 999 in increments of 1 or Null.
NumberOfShots
For each member of SamplesIn, specifies the number of redundant times the sample is illuminated with the excitation laser at each point in the SinglePoint, Grid, Rings, or Coordinates SamplingPattern.
Default Calculation: If SamplingPattern -> (SinglePoint, Grid, Rings, or Coordinates), NumberOfShots will be set to 5.
Pattern Description: Greater than or equal to 1 and less than or equal to 9999 in increments of 1 or Null.
SamplingCoordinates
For each member of SamplesIn, specifies positions at which to record spectra. The coordinates are referenced to the center of the well, which has coordinates of (0,0,0).
Default Calculation: If the SamplingPattern is set to Custom, SamplingCoordinates will be set to randomly sample 20 points in the well with the Z-dimension of 0.
Programmatic Pattern: ({{RangeP[-125*Millimeter, 125*Millimeter], RangeP[-85*Millimeter, 85*Millimeter], RangeP[-10*Millimeter, 10*Millimeter]}..} | Automatic) | Null
Protocol
NumberOfReplicates
The number of times to repeat spectroscopy reading on each provided sample. If Aliquot -> True, this also indicates the number of times each provided sample will be aliquoted.
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and when the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise set to Model[Container, Vessel, "2mL Tube"].
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSamplePreparation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentratedBufferDilutionFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Warnings and Errors
Messages (52)
ExcessiveRamanSamplingTime (1)
IncompatibleRamanSampleTypes (1)
IncorrectRamanSampleType (1)
InstrumentPrecision (1)
InvalidRamanTabletProcessingRequested (1)
LongRamanSamplingTime (1)
MissingNumberOfShots (1)
MissingRamanAdjustmentEmissionWavelength (1)
MissingRamanAdjustmentTarget (1)
MissingRamanMicroscopeImageExposureTime (1)
MissingRamanMicroscopeImageLightIntensity (1)
MissingRamanSamplingCoordinates (1)
MissingRamanSamplingDimension (1)
MissingRamanTabletProcessing (1)
NoRamanAdjustmentEmissionWavelengthRequired (1)
NoRamanAdjustmentSampleRequired (1)
NoRamanAdjustmentTargetRequired (1)
ObjectDoesNotExist (4)
RamanMaxSpeedExceeded (1)
RamanMissingFilledSquareNumberOfTurns (1)
RamanMissingNumberOfSamplingPoints (1)
RamanMissingSpiralFillArea (1)
RamanMissingSpiralInnerDiameter (1)
RamanMissingSpiralOuterDiameter (1)
RamanMissingSpiralResolution (1)
RamanNotEnoughSample (1)
RamanObjectiveMisMatch (1)
RamanSampleTypeRequiresDissolution (1)
RamanSampleWithoutBlank (1)
RamanSamplingCoordinatesPrecision (1)
RamanSamplingPatternOutOfBounds (1)
RamanSwappedInnerOuterDiameter (1)
RamanSwappedXDimensionStepSize (1)
RamanSwappedYDimensionStepSize (1)
RamanSwappedZDimensionStepSize (1)
RamanTabletProcessingInconsistancy (1)
TooManyRamanSampleInputs (1)
UnusedRamanMicroscopeImageExposureTime (1)
UnusedRamanMicroscopeImageLightIntensity (1)
UnusedRamanSamplingPatternParameterOption (1)
Possible Issues
Uneven Surface
Uneven surfaces on processed tablets may not result in reliable spectra depending when sampling at a fixed z-dimension. Use TabletProcessing -> Crush to measure the tablet as a powder or select a SamplingPattern that allows adjustment along the Z-axis, such as Coordinates or Grid to analyze the sample composition at multiple depths.
Background Fluorescence
Strongly fluorescent samples will be often result in broad background features which overwhelm the weaker Raman scattering. Post processing techniques can generally be used to remove these features. If the fluorescence arises from an impurity or solvent used to dilute the sample, consider purifying the sample or utilizing a different solvent.
Weak Signal
Set LaserPower and ExposureTime to Optimize to manually adjust the power and exposure settings for your sample. Increase the LaserPower or Exposure manually, keeping in mind that high power can lead to sample damage and detector saturation will impact spectral resolution. Conversely, LaserPower under 20% might not generate any signal.
Sample Damage
Sample damage may be evidenced by a broadening of sample peaks over the course of the measurement or changes in the Raman spectrum between repeated measurements. Decrease the LaserPower and increase Exposure to compensate. If the sample is darkly colored it is possible that laser damage is inevitable in which case it is best to use a SamplingPattern to prevent repeated measurement of damaged areas. A solution of the sample may also help to prevent heating and damage.
Background Overwhelms Signal
Background signal from the solvent, matrix, or glass sample plate may convolute weak Raman scattering peaks. If this is problematic, consider setting a Blank sample.
Cosmic Radiation
Last modified on Mon 8 Sep 2025 11:11:17