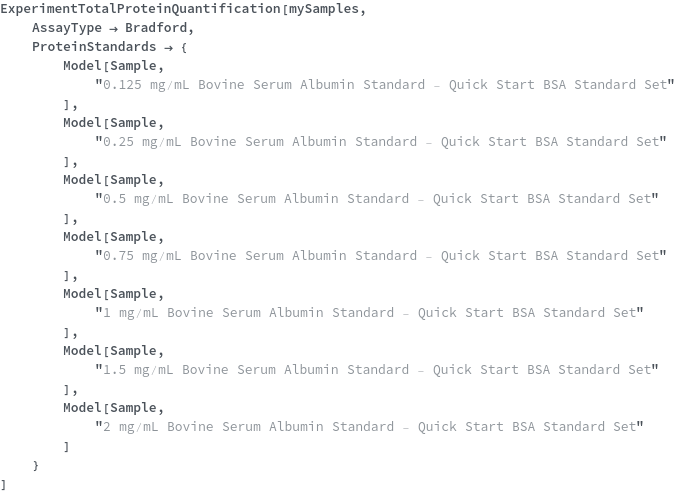
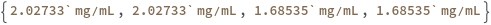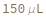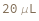ExperimentTotalProteinQuantification
ExperimentTotalProteinQuantification[Samples]⟹Protocol
generates a Protocol object for running an absorbance- or fluorescence-based assay to determine the total protein concentration of input Samples.
The quantification of protein concentration in solution is an analytical biochemical technique which most often uses spectroscopy. ExperimentTotalProteinQuantification supports three such assays: the Bradford assay, the bicinchoninic acid (BCA) assay, and the Quant-iT fluorescence quantification assay. In all three assays, standards of known protein concentrations and samples are each mixed with a fixed volume of a quantification reagent. In the absorbance-based assays, Bradford and BCA, the absorbance reading at a specific wavelength correlates to the solution’s protein concentration. In the fluorescence-based assay, the fluorescence reading at specific excitation and emission wavelengths correlates to the protein concentration. The protein concentration values obtained from the standard curve are used to calculate the protein concentrations of the input samples.
Experimental Principles
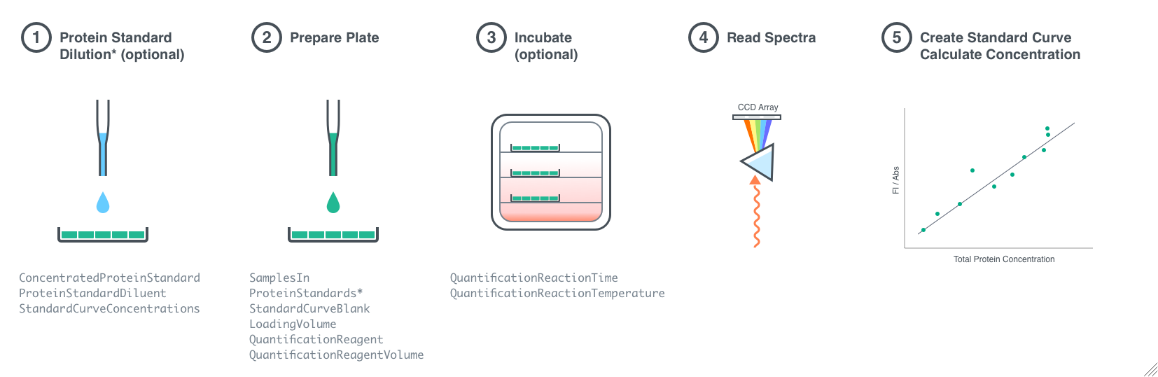
Figure 1.1: Procedural overview of a TotalProteinQuantification experiment. Step 1 (optional): A robotic liquid handler is used to dilute the ConcentratedProteinStandard with the ProteinStandardDiluent to the StandardCurveConcentrations in the StandardDilutionPlate. Step 2: A robotic liquid handler is used to mix each of the SamplesIn, StandardCurveBlank, and ProteinStandards (or contents of the StandardDilutionPlate) with the QuantificationReagent in the QuantificationPlate. Step 3 (optional): The QuantificationPlate is incubated at the QuantificationReactionTemperature for a duration of QuantificationReactionTime. Step 4: The QuantificationPlate is loaded into the appropriate plate reader. Step5: The absorbance or fluorescence spectra are recorded for each well of the QuantificationPlate. Step 6: Absorbance or fluorescence spectra from the standard protein samples are used to create a standard curve, and the protein concentrations of the input samples are calculated from this curve.
Instrumentation
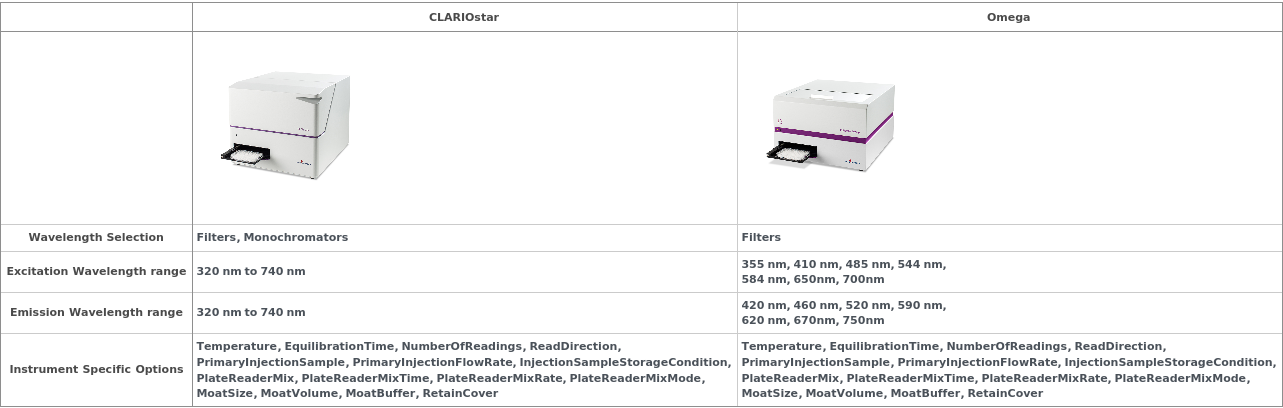
FLUOstar Omega
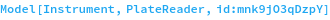
Figure 2.1.1: The Omega is a filter-based instrument. It has two independently spinning filter wheels which can be used to support an arbitrary combination of excitation and emission wavelengths. It has eight filter slots in each wheel and all 8 of these filters can be used in a single experiment - though reading will be sequential.
The plate chamber can be headed up to 45°C and it can mix the plate at up to 700 RPM before and/or during the run.
The reader has two 300μL syringe pump injectors which can be used for 0.5 - 300 μL injections of two unique samples at up to 4 time points during the run.
There are two primary kinetics read modes. It can perform all readings, injections and mixing for a single well before moving onto the next well or it can read in cycles, reading all the assay wells in the plate again and again until the RunTime has been reached. The second mode is recommended for most assays, except those that use very fast kinetics.
CLARIOstar
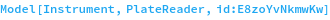
Figure 2.2.1: The CLARIOstar is primarily a monochromator-based plate reader and can thus support arbitrary combinations of excitation and emission wavelengths. Up to five wavelength pairs can be read in a single run. The CLARIOstar also has specialty excitation and emission filters which be used for particularly sensitive assays, including TR-FRET.
The plate chamber can be heated up to 45°C and it can mix the plate at up to 700 RPM before and/or during the run.
The reader has two 300μL syringe pump injectors which can be used for 0.5 - 300 μL injections of two unique samples at up to 4 time points during the run.
There are two primary kinetics read modes. It can perform all readings, injections and mixing for a single well before moving onto the next well or it can read in cycles, reading all the assay wells in the plate again and again until the RunTime has been reached. The second mode is generally recommended for assays except those that use very fast kinetics.
Experiment Options
General
AssayType
The style of protein quantification assay that is run in this experiment. The Bradford assay is an absorbance-based assay that relates changes in Coomassie Brilliant Blue G-250 absorbance at the QuantificationWavelength to protein concentration. The BCA (Smith bicinchoninic acid) assay is an absorbance-based assay which relates changes in the absorbance of a copper-BCA complex at the QuantificationWavelength to protein concentration. The FluorescenceQuantification assay relates increase in fluorescence of the QuantificationReagent at the QuantificationWavelength to protein concentration. For more information about compatible reagents and troubleshooting of the three default assays, please see Object[Report,Literature,"Quick Start Bradford Protein Assay Instruction Manual"], Object[Report,Literature,"Pierce BCA Protein Assay Kit Instructions"], and Object[Report, Literature, "Quant-iT Protein Assay Kit User Guide"].
Default Calculation: The AssayType is set to be Bradford if specified QuantificationReagent is or has a Model of Model[Sample, "Quick Start Bradford 1x Dye Reagent"], to be BCA if the QuantificationReagent is or has a Model of Model[Sample,StockSolution,"Pierce BCA Protein Assay Quantification Reagent"], to be FluorescenceQuantification if the QuantificationReagent is or has a Model of Model[Sample,StockSolution,"Quant-iT Protein Assay Quantification Reagent"], or to be Custom if the QuantificationReagent is of another Model or is an Object with no Model. If the QuantificationReagent is not specified, the AssayType is set to be Bradford if the DetectionMode is or resolves to Absorbance, or to FluorescenceQuantification if the DetectionMode is or resolves to Fluorescence.
DetectionMode
The physical phenomenon that is observed as the source of the signal for protein quantification in this experiment.
Default Calculation: The DetectionMode is set to be Fluorescence if the AssayType is specified as FluorescenceQuantification or the ExcitationWavelength or Emission-related Options are set. Otherwise, the DetectionMode is set to be Absorbance.
Instrument
The plate reader that is used to measure the absorbance or fluorescence of the mixture of QuantificationReagent and either the input samples or ProteinStandards (or diluted ConcentratedProteinStandard) present in the QuantificationPlate.
Default Calculation: The Instrument is set to be Model[Instrument, PlateReader, "FLUOstar Omega"] if DetectionMode is Absorbance, or to be Model[Instrument,PlateReader,"CLARIOstar"] if DetectionMode is Fluorescence.
Pattern Description: An object of type or subtype Model[Instrument, PlateReader] or Object[Instrument, PlateReader]
Programmatic Pattern: ObjectP[{Model[Instrument, PlateReader], Object[Instrument, PlateReader]}] | Automatic
NumberOfReplicates
The number of wells of the QuantificationPlate in which each sample is mixed with the QuantificationReagent. The protein concentration of the input sample is determined by the average absorbance or fluorescence value of the replicate samples. When the Aliquot and NumberOfReplicates options are both set, one Aliquot is performed on each input sample, and the SampleVolume is transferred from each AliquotSample into NumberOfReplicates wells of the QuantificationPlate.
Standard Curve
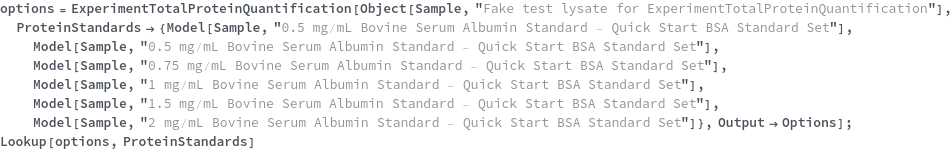
ProteinStandards
The solutions of standard protein that are mixed with the QuantificationReagent to create the standard curve. The standard concentration curve plotting Absorbance or Fluorescence versus mass concentration is used to determine the protein concentration of the input samples. If specified, the ConcentratedProteinStandard, StandardCurveConcentrations, and ProteinStandardDiluent options must not be specified.
Default Calculation: If any of the ConcentratedProteinStandard, StandardCurveConcentrations or ProteinStandardDiluent options are specified, the ProteinStandards is set to Null. If the DetectionMode is Fluorescence, the ProteinStandards is set to be {Model[Sample,"25 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"],Model[Sample,"50 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"],Model[Sample,"100 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"],Model[Sample,"200 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"],Model[Sample,"300 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"],Model[Sample,"400 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"],Model[Sample,"500 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"]}. If the DetectionMode is Absorbance, the ProteinStandards is set to be {Model[Sample,"0.125 mg/mL Bovine Serum Albumin Standard - Quick Start BSA Standard Set"],Model[Sample,"0.25 mg/mL Bovine Serum Albumin Standard - Quick Start BSA Standard Set"],Model[Sample,"0.5 mg/mL Bovine Serum Albumin Standard - Quick Start BSA Standard Set"],Model[Sample,"0.75 mg/mL Bovine Serum Albumin Standard - Quick Start BSA Standard Set"],Model[Sample,"1 mg/mL Bovine Serum Albumin Standard - Quick Start BSA Standard Set"],Model[Sample,"1.5 mg/mL Bovine Serum Albumin Standard - Quick Start BSA Standard Set"],Model[Sample,"2 mg/mL Bovine Serum Albumin Standard - Quick Start BSA Standard Set"]}.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ConcentratedProteinStandard
The concentrated solution of standard protein that is diluted with the ProteinStandardDiluent to the StandardCurveConcentrations to create the standard curve. If ConcentratedProteinStandard is specified, the ProteinStandards option must not be specified.
Default Calculation: If the ProteinStandards option is specified, the ConcentratedProteinStandard is set to be Null. Otherwise, it is set to be Model[Sample,"2 mg/mL Bovine Serum Albumin Standard"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
StandardCurveConcentrations
The mass concentrations that the ConcentratedProteinStandard is diluted to with the ProteinStandardDiluent to create the standard curve. The standard concentration curve plotting Absorbance or Fluorescence versus mass concentration is used to determine the protein concentration of the input samples. If StandardCurveConcentrations is specified, the ProteinStandards option must not be specified.
Default Calculation: If the ProteinStandards option is specified, the StandardCurveConcentrations is set to be Null. The option is set to be {0.025 mg/mL,0.05 mg/mL,0.1 mg/mL,0.2 mg/mL,0.3 mg/mL,0.4 mg/mL,0.5 mg/mL} if the DetectionMode is Fluorescence and to {0.125 mg/mL,0.25 mg/mL,0.5 mg/mL,0.75 mg/mL,1 mg/mL,1.5 mg/mL,2 mg/mL} otherwise.
Pattern Description: List of one or more greater than or equal to 0.001 milligrams per milliliter and less than or equal to 2 milligrams per milliliter entries or Null.
Programmatic Pattern: ({RangeP[0.001*(Milligram/Milliliter), 2*(Milligram/Milliliter)]..} | Automatic) | Null
ProteinStandardDiluent
The sample that is used to dilute the ConcentratedProteinStandard to the StandardCurveConcentrations. If ProteinStandardDiluent is specified, the ProteinStandards option must not be specified.
Default Calculation: If the ProteinStandards option is specified, the ProteinStandardDiluent is set to be Null. Otherwise, it is set to be Model[Sample, "Milli-Q water"].
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
StandardCurveBlank
Default Calculation: The StandardCurveBlank is set to be the ProteinStandardDiluent if the ProteinStandardDiluent, ConcentratedProteinStandard, or StandardCurveConcentrations options are specified, to Model[Sample, "Milli-Q water"] if none of these three options are specified and the DetectionMode is Absorbance, or to Model[Sample,"0 ng/uL Bovine Serum Albumin Standard - Quant-iT Protein Assay Kit"] if none of these three options are specified and the DetectionMode is Fluorescence.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
StandardCurveReplicates
The number of wells to add each concentration of ProteinStandards (or diluted ConcentratedProteinStandard) to in the QuantificationPlate. The standard curve is calculated by averaging the absorbance or fluorescence values of the protein standards at each concentration and plotting that average value versus the MassConcentration.
Post Experiment
ProteinStandardsStorageCondition
The non-default condition under which the ProteinStandards of this experiment should be stored. By default, ProteinStandard will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
ConcentratedProteinStandardStorageCondition
The non-default condition under which the ConcentratedProteinStandard of this experiment should be stored. By default, ConcentratedProteinStandard will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Quantification
LoadingVolume
The amount of each input sample, ProteinStandard, or diluted ConcentratedProteinStandard that is mixed with the QuantificationReagent in the QuantificationPlate before the absorbance or fluorescence of the mixture at the QuantificationWavelength is determined.
Default Calculation: The LoadingVolume is set to 10 uL if the DetectionMode is Fluorescence, 25 uL if the AssayType is BCA, or 5 uL is the AssayType is Bradford or Null.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 150 microliters.
QuantificationReagent
The sample that is added to the input samples and ProteinStandards or dilutions of the ConcentratedProteinStandard in the QuantificationPlate. The QuantificationReagent undergoes a change in absorbance and/or fluorescence in the presence of proteins. This change in absorbance or fluorescence is used to quantify the amount of protein present in the input samples and ProteinStandards.
Default Calculation: The QuantificationReagent is set to the Quant-iT stock solution if DetectionMode is Fluorescence, to Model[Sample,"Quick Start Bradford 1x Dye Reagent"] if AssayType is Bradford or Null, and to BCA reagent stock solution if AssayType is BCA.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
QuantificationReagentVolume
The amount of the QuantificationReagent that is added to each appropriate well of the QuantificationPlate. The QuantificationReagent is mixed with either the input samples or the ProteinStandards (or the diluted ConcentratedProteinStandard).
Default Calculation: The QuantificationReagentVolume is automatically set to 200 uL if the DetectionMode is Fluorescence or the AssayType is BCA, or 250 uL if the AssayType is Bradford or Null.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 299 microliters.
QuantificationReactionTime
The duration which the mixtures of QuantificationReagent and input samples or ProteinStandards (or diluted ConcentratedProteinStandard) are heated to QuantificationReactionTemperature on a heat block before the absorbance or fluorescence of the mixture is measured.
Default Calculation: The QuantificationReactionTime is set to 1 hour if the AssayType is BCA, to 5 minutes if the AssayType is not BCA but the QuantificationReactionTemperature is specified, and to Null otherwise.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 180 minutes or Null.
QuantificationReactionTemperature
The temperature which the mixtures of QuantificationReagent and input samples or ProteinStandards (or diluted ConcentratedProteinStandard) are heated to on a heat block before the absorbance or fluorescence of the mixture is measured.
Default Calculation: The QuantificationReactionTemperature is set to 25 Celsius if the QuantificationReactionTime has been set or has resolved to a non-Null value, and to Null otherwise.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 60 degrees Celsius or Null.
ExcitationWavelength
The wavelength of light at which the protein-bound QuantificationReagent is excited when DetectionMode is set to Fluorescence.
Default Calculation: The ExcitationWavelength is set to be Null if the DetectionMode is Absorbance, or to 470 nm otherwise.
Pattern Description: Greater than or equal to 320 nanometers and less than or equal to 740 nanometers or Null.
QuantificationWavelength
The wavelength(s) at which quantification analysis is performed to determine concentration. If multiple wavelengths are specified, the absorbance or fluorescence values from the wavelengths are averaged for standard curve determination and quantification analysis.
Default Calculation: The QuantificationWavelength is set to be 570 nm if the DetectionMode is Fluorescence, to 595 nm if AssayType is Bradford or Null, and to 562 nm otherwise.
Programmatic Pattern: (RangeP[320*Nanometer, 1000*Nanometer] | {RangeP[320*Nanometer, 1000*Nanometer]..} | RangeP[320*Nanometer, 1000*Nanometer, 1*Nanometer] ;; RangeP[320*Nanometer, 1000*Nanometer, 1*Nanometer]) | Automatic
QuantificationTemperature
Pattern Description: Ambient or greater than or equal to 25 degrees Celsius and less than or equal to 45 degrees Celsius.
QuantificationEquilibrationTime
The length of time for which the QuantificationPlate equilibrates at the QuantificationTemperature in the plate reader before being read.
NumberOfEmissionReadings
The number of redundant readings which should be taken by the detector to determine a single averaged fluorescence intensity reading for each wavelength.
Default Calculation: The NumberOfEmissionReadings is set to be Null if the DetectionMode is Absorbance. If the DetectionMode is Fluorescence, the NumberOfEmissionReadings is set to be 100.
Pattern Description: Greater than or equal to 1 and less than or equal to 200 in increments of 1 or Null.
EmissionReadLocation
Default Calculation: The EmissionReadLocation is set to be Null if the DetectionMode is Absorbance. If the DetectionMode is Fluorescence, the EmissionReadLocation is set to be Top.
EmissionGain
The gain which is applied to the signal reaching the primary detector during the emission scan when DetectionMode is Fluorescence, measured as a percentage of the strongest signal in the QuantificationPlate.
Default Calculation: The EmissionGain is set to be Null if the DetectionMode is Absorbance. If the DetectionMode is Fluorescence, the EmissionGain is set to be 90%.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
PreparatoryPrimitives
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more a primitive with head Define, Transfer, Mix, Aliquot, Consolidation, FillToVolume, Incubate, Filter, Wait, Centrifuge, or Resuspend entries or Null.
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentrationFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Preferred Input Containers
Warnings and Errors
Messages (34)
ConflictingStandardsStorageCondition (2)
InvalidTotalProteinConcentratedProteinStandardOptions (1)
InvalidTotalProteinFluorescenceQuantificationWavelength (1)
InvalidTotalProteinQuantificationStandardCurveOptions (1)
InvalidTotalProteinResolvedExcitationWavelength (1)
NonDefaultTotalProteinQuantificationReaction (1)
NotEnoughTotalProteinQuantificationWellsAvailable (1)
TooManyTotalProteinQuantificationInputs (1)
TotalProteinQuantificationAbsorbanceFluorescenceOptionsMismatch (1)
TotalProteinQuantificationAssayTypeDetectionModeMismatch (1)
TotalProteinQuantificationConcentratedProteinStandardInvalid (1)
TotalProteinQuantificationCustomAssayTypeInvalid (1)
TotalProteinQuantificationDuplicateProteinStandards (1)
TotalProteinQuantificationFluorescenceOptionsMismatch (1)
TotalProteinQuantificationInstrumentFluoresceneceOptionsMisMatch (1)
TotalProteinQuantificationInstrumentOptionMismatch (1)
TotalProteinQuantificationInvalidQuantificationWavelengthList (1)
TotalProteinQuantificationInvalidQuantificationWavelengthSpan (1)
TotalProteinQuantificationInvalidVolumes (1)
TotalProteinQuantificationLoadingVolumeLow (1)
TotalProteinQuantificationMultipleProteinStandardIdentityModels (1)
TotalProteinQuantificationNullFluorescenceOptionsMismatch (1)
TotalProteinQuantificationNullProteinStandardConcentration (1)
TotalProteinQuantificationQuantificationReagentVolumeLow (1)
TotalProteinQuantificationReactionOptionsMisMatch (1)
TotalProteinQuantificationReagentNotOptimal (1)
TotalProteinQuantificationTotalVolumeLow (1)
TotalProteinQuantificationUnsupportedInstrument (1)
TotalProteinQuantificationWavelengthMismatch (1)
Last modified on Mon 19 Dec 2022 16:54:27