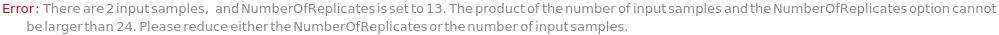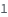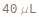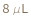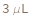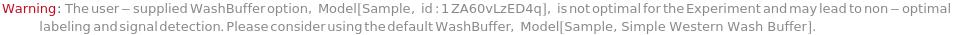ExperimentWestern
ExperimentWestern[Samples,Antibodies]⟹Protocol
generates a Protocol object for running a western blot using capillary electrophoresis.
Western blotting is an analytical molecular biology technique used to detect the presence of a specific protein in a complex mixture. ExperimentWestern replaces the traditional slab gel western blot technique with an automated capillary-based immunoassay. As in the traditional assay, analytes are separated through a gel matrix by size, then labeled with primary and secondary antibodies. The presence of secondary antibodies, and thus the analytes of interest, is detected by chemiluminescence. This technique is often used to qualitatively confirm the presence or absence of a single protein in a cell lysate, for example.
Experimental Principles
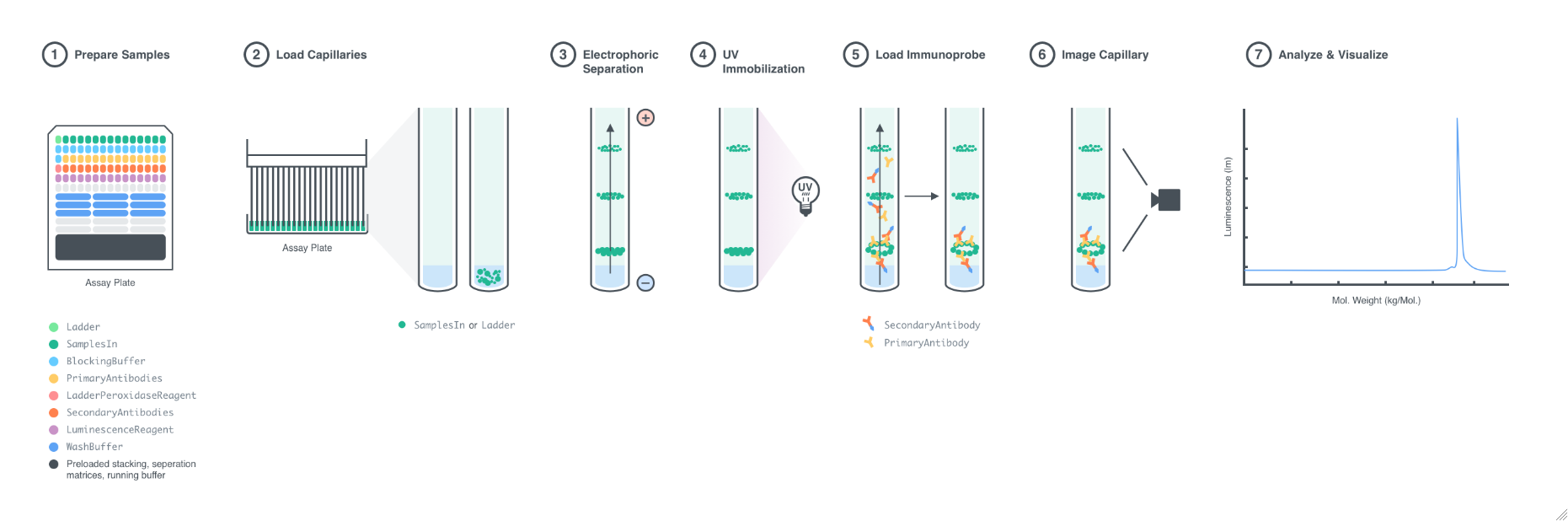
Figure 1.1: Procedural overview of a Western experiment. Step 1: PrimaryAntibodies are diluted with the PrimaryAntibodyDiluents in the AntibodyPlate. Input samples are mixed with the LoadingBuffer in the SamplePlate. The AssayPlate is loaded with the input samples, PrimaryAntibodies, BlockingBuffers, SecondaryAntibodies, LuminescenceReagent, Ladder, LadderPeroxidaseReagent, and WashBuffer. Step 2: On the Instrument, the capillaries are loaded with separating matrix, stacking matrix, and input samples. Step 3: Voltage is applied across the capillaries as the analytes in the input samples are electrophoretically separated. Step 4: The analytes are immobilized on the inside surface of the capillaries with UV light. Step 5: The capillaries are incubated with the BlockingBuffers, the PrimaryAntibodies, the SecondaryAntibodies, and the LadderPeroxidaseReagent, in order, with an incubation with the WashBuffer between each step. Step 6: The capillaries are incubated with the LuminescenceReagent, and the chemiluminescent signal is detected by a CCD camera. Step 7: The output data is an electropherogram of relative signal intensity versus molecular weight.
Instrumentation
Wes

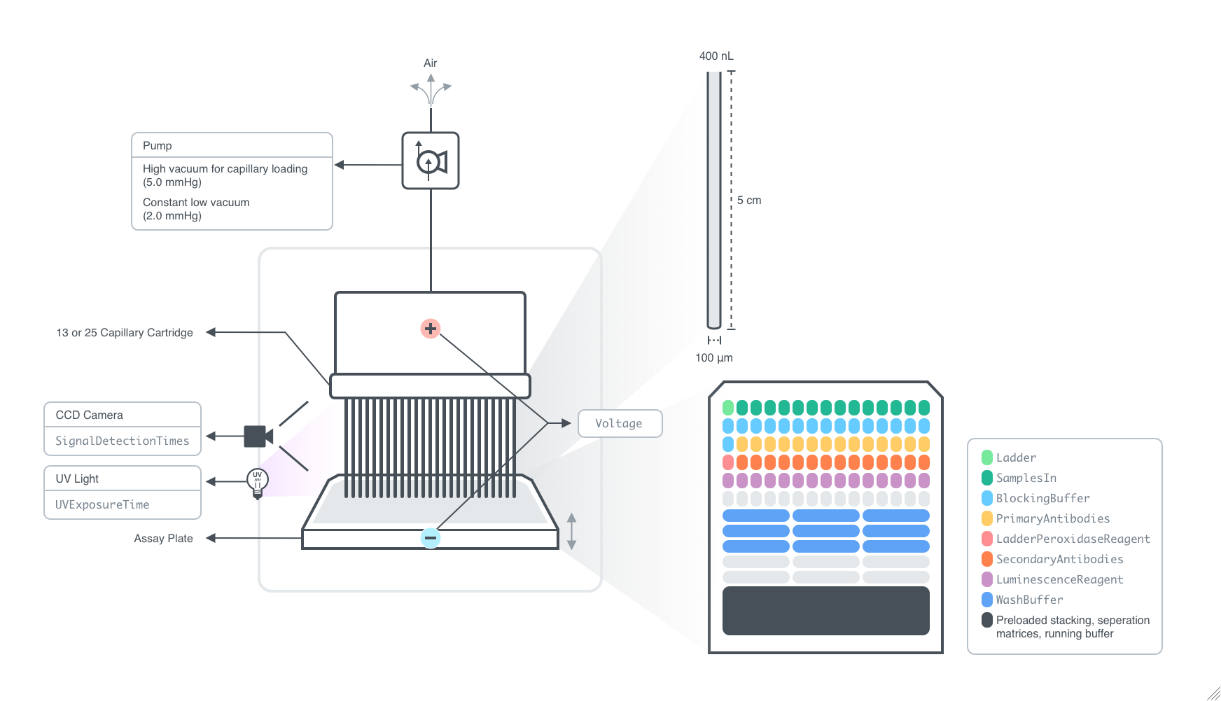
Figure 2.1.1: Instrument diagram for the Wes system: The instrument has an internal plate position and a slot for the CapillaryCartridge. The AssayPlate, containing input samples, LabelingReagent, BlockingBuffer, PeroxidaseReagent, LuminescenceReagent, WashBuffer, Ladder, LadderBlockingBuffer, and preloaded with the separation matrix, stacking matrix, and running buffer, is placed into the plate slot of the instrument. The CapillaryCartridge is placed into its slot. The instrument deck moved the AssayPlate position during the run so the capillaries can be placed into the appropriate wells. The instrument loads the various reagents into the capillaries by creating a pressure differential across the capillaries. With the bottom ends of the capillaries in the running buffer, analytes in each input sample and Ladder are electrophoretically separated as Voltage is applied across the capillaries. Chemiluminescent signal is produced from the interaction between the LuminescenceReagent and the horseradish peroxidase present on the PeroxidaseReagent. The signal is captured by a CCD camera and processed into an electropherogram for each capillary.
Experiment Options
General
MolecularWeightRange
The range of protein sizes for which good resolution can be expected in this experiment. LowMolecularWeight corresponds to 2-40 kDa, MidMolecularWeight corresponds to 12-230 kDa, and HighMolecularWeight corresponds to 66-440 kDa. This molecular weight range determines which western assay plate, the specialized plate that is inserted into the western instrument, will be used in the experiment.
Default Calculation: In ExperimentWestern, the MolecularWeightRange is automatically set to be MidMolecularWeight if the average Expected MolecularWeight of the input PrimaryAntibodies are between 12 and 120 kDa, LowMolecularweight if between 0 and 12 kDa, and HighMolecularWeight if greater than 120 kDa. In ExperimentTotalProteinDetection, the the MolecularWeightRange is automatically set depending on the Ladder option.
Instrument
The Western instrument used for this experiment. The instrument loads the input samples into the capillaries, and performs the sample separation, antibody or biotin labeling reagent incubation, and signal detection after being loaded with a specialized western assay plate containing the input samples, antibodies or biotin labeling reagent, buffers, and other detection reagents.
Pattern Description: An object of type or subtype Object[Instrument, Western] or Model[Instrument, Western]
NumberOfReplicates
The number of capillaries each input sample will be run though. For example, ExperimentFunction[{input2,input2}, NumberOfReplicates->2] is equivalent to ExperimentFunction[{input1,input1,input2,input2}].
Denaturation
Denaturing
Indicates if the mixture of input samples and prepared master mix will be denatured by heating before preparing the samples for loading. The Denaturation options control the temperature and duration of this denaturation.
Default Calculation: The Denaturing is automatically set to False if DenaturingTemperature, DenaturingTime, or Denaturant is specified to be Null. Otherwise, the option is set to True.
DenaturingTemperature
The temperature which the mixture of input samples and loading buffer will be heated to before being transferred to the western assay plate (the specialized plate that is inserted into the western instrument).
Default Calculation: The DenaturingTemperature is automatically set to 95 Celsius if Denaturing is True, and to Null if Denaturing is False.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 95 degrees Celsius or Null.
DenaturingTime
The duration which the mixture of input samples and loading buffer will be heated to DenaturingTemperature before being transferred to the western assay plate (the specialized plate that is inserted into the western instrument).
Default Calculation: The DenaturingTime is automatically set to 5 minutes if Denaturing is True, and to Null if Denaturing is False.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 120 minutes or Null.
Matrix & Sample Loading
SeparatingMatrixLoadTime
The duration for which the separating matrix will be loaded into the capillary. The Matrix and Sample Loading options determine the composition of the matrix inside the capillary, the sample preparation parameters, and the amount of sample loaded into the capillary.
StackingMatrixLoadTime
Default Calculation: The StackingMatrixLoadTime is automatically set to 15 seconds if the MolecularWeightRange is MidMolecularWeight, and to 12 seconds otherwise.
SampleVolume
The amount of each input sample that will be mixed with the LoadingBuffer before the mixture is denatured, and a portion of the mixture, the LoadingVolume, is loaded into the western assay plate (the specialized plate that is inserted into the western instrument).
Pattern Description: Greater than or equal to 7 microliters and less than or equal to 10 microliters.
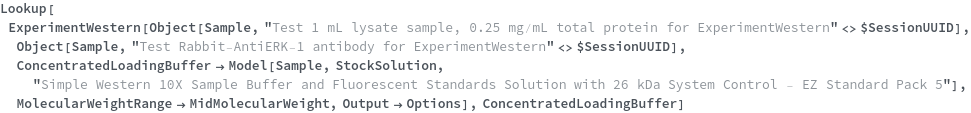
ConcentratedLoadingBuffer
The control protein and sample buffer-containing solution, provided by the instrument supplier, that will be mixed with either the Denaturant or deionized water to make the LoadingBuffer, before a portion of the mixture, the LoadingBufferVolume, is mixed with the SampleVolumes.
Default Calculation: The ConcentratedLoadingBuffer is automatically set to be a Simple Western 10X Sample Buffer and Fluorescent Standards Solution from the same kit as the Ladder if the Ladder is set. If the Ladder is not set, the ConcentratedLoadingBuffer is automatically set to be a Simple Western 10X Sample Buffer and Fluorescent Standards Solution which covers the molecular weight span of the MolecularWeightRange.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
ConcentratedLoadingBufferVolume
The amount of the ConcentratedLoadingBuffer that will be mixed with either the Denaturant or deionized water, depending on if Denaturing is True or False.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 40 microliters.
Denaturant
The solution, containing the protein denaturing agent used to denature proteins present in the input samples, that will be mixed with ConcentratedLoadingBuffer to make the LoadingBuffer.
Default Calculation: The Denaturant is automatically set to be a Model[Sample,StockSolution,"Simple Western 400 mM DTT"] from the same kit as the ConcentratedLoadingBuffer if the ConcentratedLoadingBuffer is set. Otherwise, it automatically resolves to be the 400mM DTT solution from EZ Standard Pack 1, 3 or 5, depending on the MolecularWeightRange.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
DenaturantVolume
Default Calculation: The DenaturantVolume is automatically set to be Null if the Denaturant is Null, or to 40 uL otherwise.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 40 microliters or Null.
WaterVolume
The amount of deionized water that will be mixed with the ConcentratedLoadingBuffer in lieu of the Denaturant if Denaturing is set to False.
Default Calculation: The WaterVolume is automatically set to be 40 uL if Denaturing is False, and Null otherwise.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 40 microliters or Null.
LoadingBufferVolume
The amount of prepared LoadingBuffer (with ConcentratedLoadingBuffer and either Denaturant or deionized water already added) that will be mixed with each input sample before the mixture is denatured, and a portion of the mixture, the LoadingVolume, is loaded into the western assay plate (the specialized plate that is inserted into the western instrument).
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 2.5 microliters.
LoadingVolume
The amount of each mixture of input sample and LoadingBuffer that will be loaded into the western assay plate after sample denaturation.
Pattern Description: Greater than or equal to 5 microliters and less than or equal to 9 microliters.
Ladder
The biotinylated ladder, provided by the instrument supplier, which will be used as a standard reference ladder in the Experiment. After electrophoretic separation, the ladder will be labeled with the LadderPeroxidaseReagent in ExperimentWestern or the PeroxidaseReagent in ExperimentTotalProteinDetection so that each protein band is visible during the signal detection step.
Default Calculation: The Ladder is automatically set to be a Simple Western Biotinylated Ladder Solution from the same kit as the ConcentratedLoadingBuffer if the ConcentratedLoadingBuffer is set. If the ConcentratedLoadingBuffer is not set, the Ladder is automatically set to be a Simple Western Biotinylated Ladder Solution which covers the molecular weight span of the MolecularWeightRange.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
LadderVolume
The volume of the Ladder to be aliquotted into its well in the western assay plate (the specialized plate that is inserted into the western instrument).
Pattern Description: Greater than or equal to 3 microliters and less than or equal to 10 microliters.
SampleLoadTime
Default Calculation: The SampleLoadTime is automatically set to 8 seconds if the MolecularWeightRange is HighMolecularWeight, and to 9 seconds otherwise. For more information on how the SampleLoadTime affects the experiment, please refer to Object[Report, Literature, "Simple Western Size Assay Development Guide"].
Separation & Immobilization
Voltage
The voltage applied during the electrophoretic separation step. The separation and immobilization options set the instrument parameters that control the size-dependent separation of proteins present in the input samples and ladder, and the UV light-induced crosslinking of proteins to the capillary after separation.
Default Calculation: The Voltage is automatically set to 475 volts if the MolecularWeightRange is HighMolecularWeight, and to 375 volts otherwise.
Pattern Description: Greater than or equal to 1 volt and less than or equal to 500 volts in increments of 1 volt.
SeparationTime
Default Calculation: The SeparationTime is automatically set to 1620 seconds, 1500 seconds, or 1800 seconds if the MolecularWeightRange is set to LowMolecularWeight, MidMolecularWeight, or HighMolecularWeight, respectively. For more information on how the SeparationTime affects the experiment, please refer to Object[Report, Literature, "Simple Western Size Assay Development Guide"].
UVExposureTime
The duration for which the capillary will be exposed to UV-light for protein cross-linking to the capillary.
Default Calculation: The UVExposureTime is automatically set to 150 seconds if the MolecularWeightRange is HighMolecularWeight, and to 200 seconds otherwise.
WashBuffer
The buffer that will be incubated with the capillary between blocking and labeling steps to remove excess reagents.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
WashBufferVolume
The amount of WashBuffer to be loaded into each of the 15 appropriate wells of the western assay plate (the specialized plate that is inserted into the western instrument).
Pattern Description: Greater than or equal to 250 microliters and less than or equal to 500 microliters.
Blocking
BlockingBuffer
The buffer that is incubated with the capillary after the MatrixRemovalTime. Incubation in this buffer reduces non-specific antibody binding to residual matrix present in the capillary. For more information about assay development and troubleshooting unexpected results, please refer to Object[Report, Literature, "Simple Western Size Assay Development Guide"].
Default Calculation: The BlockingBuffer is automatically set to Model[Sample,"Simple Western Milk-Free Antibody Diluent"] if the Organism of the PrimaryAntibody is Goat, and to Model[Sample,"Simple Western Antibody Diluent 2"] otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
BlockingBufferVolume
The amount of BlockingBuffer that is aliquotted into the appropriate wells of the western assay plate (the specialized plate that is inserted into the western instrument).
Pattern Description: Greater than or equal to 5 microliters and less than or equal to 20 microliters.
LadderBlockingBuffer
The buffer that is incubated with the capillary containing the Ladder during both the BlockingTime and the PrimaryIncubationTime.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
LadderBlockingBufferVolume
The amount of LadderBlockingBuffer that is aliquotted into each of the two appropriate wells of the western assay plate (the specialized plate that is inserted into the western instrument).
Pattern Description: Greater than or equal to 5 microliters and less than or equal to 20 microliters.
BlockingTime
Antibody Labeling
PrimaryAntibodyVolume
The amount of the concentrated stock solution of input PrimaryAntibody that is mixed with the PrimaryAntibodyDiluent and StandardPrimaryAntibody before a portion of the mixture, the PrimaryAntibodyLoadingVolume, is loaded into the western assay plate (the specialized plate that is inserted into the western instrument). After the analyte binding, which occurs during the PrimaryIncubationTime, the presence of the PrimaryAntibody is detected by the SecondaryAntibody.
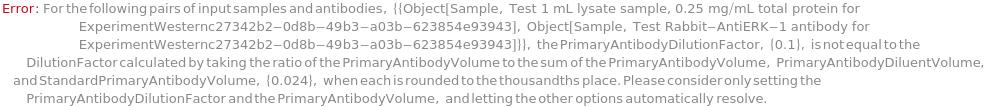
Default Calculation: The PrimaryAntibodyVolume is automatically set to 36 uL if the PrimaryAntibodyDiluent is Null. Otherwise, The option is calculated from the equation PrimaryAntibodyDilutionFactor = (PrimaryAntibodyVolume/(PrimaryAntibodyVolume + PrimaryAntibodyDiluentVolume + StandardPrimaryAntibodyVolume)) with the assumptions that The StandardPrimaryAntibodyVolume will be 10% of the total volume when SystemStandard is True, and that the total volume will be 100 uL whenever possible.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 200 microliters.
PrimaryAntibodyStorageCondition
The non-default storage condition under which the PrimaryAntibody of this experiment should be stored after the protocol is completed. If left unset, the PrimaryAntibody will be stored according to its current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
PrimaryAntibodyDiluent
The buffer which is mixed with the PrimaryAntibody to reduce the the concentration of the antibody solution that will be loaded into the western assay plate (the specialized plate that is inserted into the western instrument). For more information about assay development and troubleshooting unexpected results, please refer to Object[Report, Literature, "Simple Western Size Assay Development Guide"].
Default Calculation: The PrimaryAntibodyDiluent is automatically set to Null if the PrimaryAntibodyDilutionFactor is 1, or if the PrimaryAntibodyDiluentVolume is 0 uL or Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
PrimaryAntibodyDilutionFactor
A measure of the ratio of PrimaryAntibodyVolume to the diluted PrimaryAntibody (PrimaryAntibody plus PrimaryAntibodyDiluent plus StandardPrimaryAntibody). A PrimaryAntibodyDilutionFactor of 0.02 indicates that the diluted PrimaryAntibody solution will be comprised of 1 part concentrated PrimaryAntibody and 49 parts of either PrimaryAntibodyDiluent or a mixture of PrimaryAntibodyDiluent and StandardPrimaryAntibody. For more information about suggested primary antibody dilutions, please refer to https://www.proteinsimple.com/antibody/antibodies.html. If traditional Western Blot primary antibody dilution conditions are known, it is recommended to start with 100, 20, and 4 times the recommended traditional primary antibody dilutions for initial antibody screening. For example, if the recommended traditional Western dilution is 1:1000 (a PrimaryAntibodyDilutionFactor of 0.001), the recommended starting PrimaryAntibodyDilutionFactors would be 0.1, 0.02, and 0.004. For a new assay where Western blot parameters are not known, it is recommended to use PrimaryAntibodyDilutionFactors of 0.1, 0.02, and 0.004 for initial antibody screening. For more information about assay development and troubleshooting unexpected results, please refer to Object[Report, Literature, "Simple Western Size Assay Development Guide"].
Default Calculation: The PrimaryAntibodyDilutionFactor is automatically set to 1 if the PrimaryAntibodyDiluent is Null. If any of the PrimaryAntibodyVolume, PrimaryAntibodyDiluentVolume, or StandardPrimaryAntibodyVolume are set, the PrimaryAntibodyDilutionFactor is calculated from the equation PrimaryAntibodyDilutionFactor = (PrimaryAntibodyVolume/(PrimaryAntibodyVolume + PrimaryAntibodyDiluentVolume + StandardPrimaryAntibodyVolume)), with the assumptions that The StandardPrimaryAntibodyVolume will be 10% of the total volume when SystemStandard is True, and that the total volume will be 100 uL whenever possible. If none of the previously mentioned options are specified, the PrimaryAntibodyDilutionFactor is automatically set to the RecommendedDilution of the PrimaryAntibody.
PrimaryAntibodyDiluentVolume
The amount of the PrimaryAntibodyDiluent that is mixed with the PrimaryAntibody and the StandardPrimaryAntibody before a portion of the mixture, the PrimaryAntibodyLoadingVolume, is loaded into the western assay plate (the specialized plate that is inserted into the western instrument).
Default Calculation: The PrimaryAntibodyDiluentVolume is automatically set to Null when the PrimaryAntibodyDiluent is Null. Otherwise, The option is calculated from the equation PrimaryAntibodyDilutionFactor = (PrimaryAntibodyVolume/(PrimaryAntibodyVolume + PrimaryAntibodyDiluentVolume + StandardPrimaryAntibodyVolume)) with the assumptions that The StandardPrimaryAntibodyVolume will be 10% of the total volume when SystemStandard is True, and that the total volume will be 100 uL whenever possible.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
SystemStandard
Indicates if a StandardPrimaryAntibody and secondary antibody-HRP conjugate is used to detect a standard protein present in the ConcentratedLoadingBuffer. This option is typically set to True when trying to troubleshoot anomalous results.
StandardPrimaryAntibody
The solution containing a control antibody which detects the system control protein in the LoadingBuffer, provided by the instrument supplier, that is mixed with the PrimaryAntibody and the PrimaryAntibodyDiluent.
Default Calculation: The StandardPrimaryAntibody is automatically set to Null if SystemStandard is False. If SystemStandard is True, the option resolves to Model[Sample,"Simple Western 10X System Control Primary Antibody-Mouse"] if the Organism of the PrimaryAntibody is Mouse, and to Model[Sample,"Simple Western 10X System Control Primary Antibody-Rabbit"] otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
StandardPrimaryAntibodyStorageCondition
The non-default storage condition under which the StandardPrimaryAntibody of this experiment should be stored after the protocol is completed. If left unset, the StandardPrimaryAntibody will be stored according to its current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
StandardPrimaryAntibodyVolume
The amount of the StandardPrimaryAntibody that is mixed with the PrimaryAntibody and the PrimaryAntibodyDiluent before a portion of the mixture, the PrimaryAntibodyLoadingVolume, is loaded into the western assay plate (the specialized plate that is inserted into the western instrument).
Default Calculation: The StandardPrimaryAntibodyVolume is automatically set to Null if the SystemStandard is False. If the SystemStandard is True and the PrimaryAntibodyDilutionFactor is 1, the StandardPrimaryAntibodyVolume ia automatically set to be 10% of the PrimaryAntibodyVolume. Otherwise, the option is calculated from the equation PrimaryAntibodyDilutionFactor = (PrimaryAntibodyVolume/(PrimaryAntibodyVolume + PrimaryAntibodyDiluentVolume + StandardPrimaryAntibodyVolume)).
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 30 microliters or Null.
PrimaryAntibodyLoadingVolume
The amount of each mixture of PrimaryAntibody, PrimaryAntibodyDiluent, and StandardPrimaryAntibody that is loaded into the appropriate wells of the western assay plate (the specialized plate that is inserted into the western instrument).
Default Calculation: The PrimaryAntibodyLoadingVolume is automatically set to 10 uL, unless any input sample's total of PrimaryAntibodyVolume, PrimaryAntibodyDiluentVolume, and StandardPrimaryAntibodyVolume is less than 10 uL, in which case the option is set to the lowest such volume.
Pattern Description: Greater than or equal to 5 microliters and less than or equal to 20 microliters.
PrimaryIncubationTime
SecondaryAntibody
The antibody-HRP conjugate solution that is used to detect the PrimaryAntibody. Corresponding SecondaryAntibodies will be automatically set for any PrimaryAntibodies that are derived from Mouse, Rabbit, Human, or Goat. For PrimaryAntibodies derived from other species, the user should provide a corresponding SecondaryAntibody. For more information about assay development and troubleshooting unexpected results, please refer to Object[Report, Literature, "Simple Western Size Assay Development Guide"].
Default Calculation: The SecondaryAntibody is automatically set to Model[Sample,"Simple Western Goat-AntiRabbit-HRP"] if the Organism of the PrimaryAntibody is Rabbit, to Model[Sample,"Simple Western Goat-AntiMouse-HRP"] if Mouse, to Model[Sample,"Simple Western Goat-AntiHuman-IgG-HRP"] if Human, to Model[Sample,"Simple Western Donkey-AntiGoat-HRP"] if Goat, and to Model[Sample,"Simple Western Antibody Diluent 2"] otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
SecondaryAntibodyStorageCondition
The non-default storage condition under which the SecondaryAntibody of this experiment should be stored after the protocol is completed. If left unset, the SecondaryAntibody will be stored according to its current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
SecondaryAntibodyVolume
The amount of SecondaryAntibody solution that is aliquotted into the appropriate wells of the western assay plate (the specialized plate that is inserted into the western instrument).
Default Calculation: The SecondaryAntibodyVolume is automatically set to 10 uL if the StandardSecondaryAntibody is Null, to 9.5 uL if the StandardSecondaryAntibody is not Null but the StandardSecondaryAntibodyVolume is left as Automatic, and to 19 times the StandardSecondaryAntibodyVolume otherwise.
Pattern Description: Greater than or equal to 5 microliters and less than or equal to 19 microliters.
StandardSecondaryAntibody
The concentrated solution containing a control antibody-HRP conjugate which detects the StandardPrimaryAntibody. The StandardSecondaryAntibody is mixed with the SecondaryAntibody in cases where the PrimaryAntibody and StandardPrimaryAntibody are not derived from the same mammal - when the PrimaryAntibody is human or goat-derived.
Default Calculation: The StandardSecondaryAntibody is automatically set to Null if the StandardPrimaryAntibody is Null. If the StandardPrimaryAntibody is Model[Sample,"Simple Western 10X System Control Primary Antibody-Mouse"] and the Organism of the PrimaryAntibody is Mouse, or the StandardPrimaryAntibody is Model[Sample,"Simple Western 10X System Control Primary Antibody-Rabbit"] and the Organism of the PrimaryAntibody is Rabbit, the StandardSecondaryAntibody resolves to Null. Otherwise, the option resolves to Model[Sample, "Simple Western 20X Goat-AntiRabbit-HRP"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
StandardSecondaryAntibodyStorageCondition
The non-default storage condition under which the StandardSecondaryAntibody of this experiment should be stored after the protocol is completed. If left unset, the StandardSecondaryAntibody will be stored according to its current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
StandardSecondaryAntibodyVolume
The amount of the concentrated solution of the StandardSecondaryAntibody that is mixed with the SecondaryAntibody in cases where the PrimaryAntibody and StandardPrimaryAntibody are not derived from the same mammal - when the PrimaryAntibody is human or goat-derived.
Default Calculation: The StandardSecondaryAntibodyVolume is automatically set to Null if the StandardSecondaryAntibody is Null, and to the SecondaryAntibodyVolume divided by 19 otherwise.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 1 microliter or Null.
SecondaryIncubationTime
The duration for which the capillary is incubated with the SecondaryAntibody or the LadderPeroxidaseReagent (for the capillary containing the Ladder).
LadderPeroxidaseReagent
The sample or model of streptavidin-HRP solution which binds to the biotinylated ladder provided by the instrument supplier.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
LadderPeroxidaseReagentStorageCondition
The non-default storage condition under which the LadderPeroxidaseReagent of this experiment should be stored after the protocol is completed. If left unset, the LadderPeroxidaseReagent will be stored according to its current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
LadderPeroxidaseReagentVolume
The amount of streptavidin-HRP conjugate solution that is added to the appropriate well of the western assay plate (the specialized plate that is inserted into the western instrument).
Pattern Description: Greater than or equal to 5 microliters and less than or equal to 20 microliters.
Imaging
LuminescenceReagent
The solution, which defaults to a mixture of luminol and peroxide, that reacts with the horseradish peroxidase (HRP) attached to the SecondaryAntibody in ExperimentWestern or the PeroxidaseReagent in ExperimentTotalProteinLabeling to give off chemiluminesence which is observed during the SignalDetectionTimes.
Default Calculation: In ExperimentWestern, the LuminescenceReagent is automatically set to Model[Sample,StockSolution,"SimpleWestern Luminescence Reagent"]. In ExperimentTotalProteinDetection, the LuminescenceReagent is automatically set to Model[Sample,StockSolution,"SimpleWestern Luminescence Reagent - Total Protein Kit"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
LuminescenceReagentVolume
The amount of the LuminescenceReagent that will be aliquotted into each of the 25 appropriate wells of the western assay plate (the specialized plate that is inserted into the western instrument). The LuminescenceReagent reacts with the horseradish peroxidase (HRP) attached to the SecondaryAntibody in ExperimentWestern or the PeroxidaseReagent in ExperimentTotalProteinLabeling to give off chemiluminesence which is observed during the SignalDetectionTimes.
Pattern Description: Greater than or equal to 5 microliters and less than or equal to 15 microliters.
SignalDetectionTimes
The list of (9) exposure times in seconds for chemiluminescent signal. It is highly recommended to leave the SignalDetectionTimes as the default times, as these are the only times from which the High Dynamic Range (HDR) Detection Profile can be calculated. The HDR Detection Profile increases the dynamic range of the experiment by two orders of magnitude.
Default Value: {1 second, 2 seconds, 4 seconds, 8 seconds, 16 seconds, 32 seconds, 64 seconds, 128 seconds, 512 seconds}
Pattern Description: {First Exposure, Second Exposure, Third Exposure, Fourth Exposure, Fifth Exposure, Sixth Exposure, Seventh Exposure, Eighth Exposure, Ninth Exposure}
Programmatic Pattern: {RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second], RangeP[1*Second, 512*Second]}
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
PreparatoryPrimitives
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more a primitive with head Define, Transfer, Mix, Aliquot, Consolidation, FillToVolume, Incubate, Filter, Wait, Centrifuge, or Resuspend entries or Null.
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentrationFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Warnings and Errors
Messages (32)
ConflictingWesDenaturingOptions (1)
ConflictingWesNullOptions (1)
ConflictingWesternPrimaryAntibodyDilutionFactorOptions (1)
DiscardedSamples (2)
DuplicateName (1)
InvalidWesternDilutedPrimaryAntibodyVolume (1)
NonIdealWesternBlockingBuffer (1)
NonIdealWesternPrimaryAntibodyDiluent (1)
NonIdealWesternSecondaryAntibody (1)
NonIdealWesternStandardPrimaryAntibody (1)
NonIdealWesternStandardSecondaryAntibody (1)
NonOptimalUserSuppliedWesMolecularWeightRange (1)
NotEnoughWesLoadingBuffer (1)
NumberOfReplicatesTooHighForNumberOfWesInputSamples (1)
ObjectDoesNotExist (1)
WesConcentratedLoadingBufferNotOptimal (1)
WesConflictingStandardPrimaryAntibodyStorageOptions (1)
WesConflictingStandardSecondaryAntibodyStorageOptions (1)
WesHighDynamicRangeImagingNotPossible (1)
WesInputsShouldBeDiluted (1)
WesLadderNotOptimalForMolecularWeightRange (1)
WesOptionVolumeTooLow (1)
WesternLoadingVolumeTooLarge (1)
WesternPrimaryAntibodyVolumeLarge (1)
WesternSecondaryAntibodyVolumeLow (1)
WesternStandardPrimaryAntibodyVolumeRatioNonIdeal (1)
WesTotalProteinConcentrationNotInformed (1)
Possible Issues
Overloading
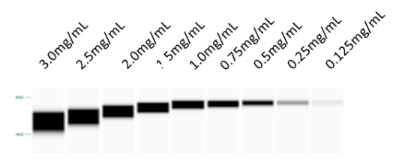
Effect of sample overloading on sizing accuracy. The maximum recommended TotalProteinConcentration for input lysate samples is between 2 and 3 mg/mL. It is recommended to dilute concentrated lysate inputs with Model[Sample, StockSolution, "Simple Western 0.1X Sample Buffer"] using the Aliquot-related sample preparation options. Please see the AssayBuffer option for an example. For novel assays, it is recommended to start with lysate concentrations of 1 and 0.25 mg/mL. These will become 0.8 and 0.2 mg/mL after the LoadingBuffer is added to input lysate samples. For "no lysate" controls, use a sample of Model[Sample, StockSolution, "Simple Western 0.1X Sample Buffer"] in place of input lysate. In the example below, lanes with overloaded samples (lanes 1-4) suffer from band size inaccuracy: bands are visibly lower than lanes 5-9.
Non-Specific Labeling
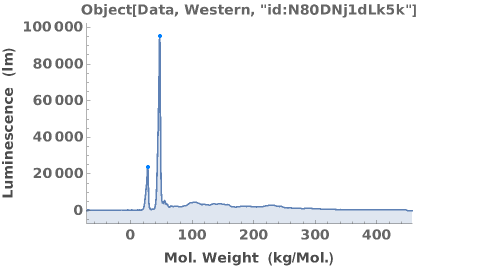
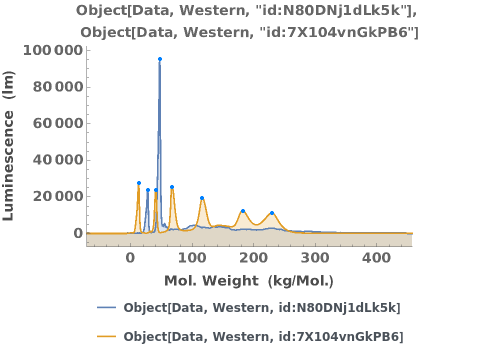
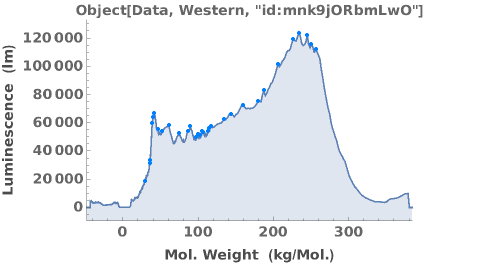
Some antibodies can bind non-specifically to the capillary. A list of antibodies and recommended dilutions that have been successfully used in capillary-based western experiments can be found at https://www.proteinsimple.com/antibody/antibodies.html. For more information about assay development and troubleshooting unexpected results, please refer to Object[Report, Literature, "Simple Western Size Assay Development Guide"]. Example data below shows multiple peaks, indicating the antibody may be non-specific.
Last modified on Thu 15 Dec 2022 12:05:44