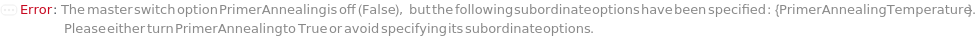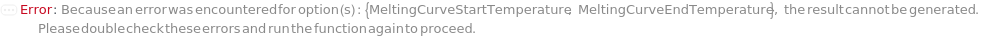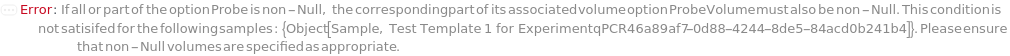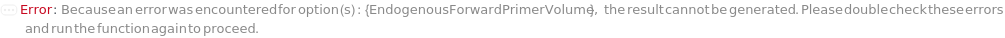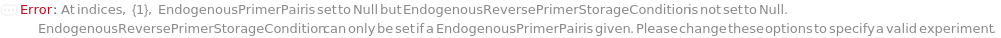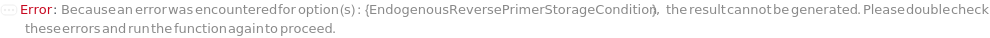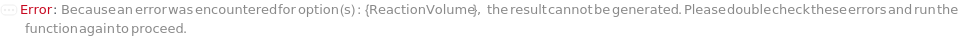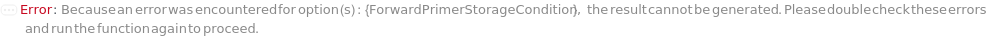General
Instrument
The instrument for running the quantitative polymerase chain reaction (qPCR) experiment.
Pattern Description: An object of type or subtype Model[Instrument, Thermocycler] or Object[Instrument, Thermocycler]
Programmatic Pattern: ObjectP[{Model[Instrument, Thermocycler], Object[Instrument, Thermocycler]}] | Automatic
ReactionVolume
The total volume of the reaction including the sample, primers, probes, dyes and master mix. If using an array card, the final reaction volume in the microfluidic well following centrifugation.
Default Calculation: Automatically set to 2 Microliter if using an array card, or 20 Microliter otherwise.
Pattern Description: Greater than or equal to 2 microliters and less than or equal to 50 microliters.
Programmatic Pattern: RangeP[2*Microliter, 50*Microliter] | Automatic
MasterMix
The stock solution composed of enzymes (DNA polymerase and optionally reverse transcriptase), buffer, dyes and nucleotides used to amplify cDNA or gDNA.
Default Calculation: Automatically set based on DuplexStainingDye and ReverseTranscription.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
MasterMixVolume
The volume of the master mix to add to the reaction. If using an array card, the initial volume of the master mix to be mixed with the sample then loaded into the array card reservoir.
Default Calculation: Automatically set based on ReactionVolume and MasterMix.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters.
Programmatic Pattern: RangeP[0.5*Microliter, 50*Microliter] | Automatic
StandardVolume
The volume of the standard to add to the reaction.
Default Calculation: Resolved automatically to 2 Microliter if a Standard is included in the experiment.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Buffer
The chemical or stock solution used to bring each reaction to ReactionVolume once all other components have been added.
Default Calculation: Automatically set to Model[Sample,"Milli-Q water"] if after the addition of all reaction components the volume has not reached ReactionVolume, or Null otherwise.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
SampleVolume
The volume of the sample from which the target will be amplified to add to the reaction. If using an array card, the initial volume of the sample to be mixed with the master mix then loaded into the array card reservoir.
Default Calculation: Automatically set to 48 Microliter if using an array card, or 2 Microliter otherwise.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters.
Programmatic Pattern: RangeP[0.5*Microliter, 50*Microliter] | Automatic
Index Matches to: experiment samples
BufferVolume
The volume, if any, of buffer added to bring the total volume up to ReactionVolume (if using an array card, ReactionVolume*48).
Default Calculation: Automatically set to a value based on the volume of all other reaction components.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters.
Programmatic Pattern: RangeP[0.5*Microliter, 50*Microliter] | Automatic
Index Matches to: experiment samples
ForwardPrimerVolume
The total volume of each forward primer to add to the reaction.
Default Calculation: Automatically set to Null if ArrayCard is True, otherwise it is set to ReversePrimerVolume (if specified) or 1 Microliter.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or list of one or more greater than or equal to 0.5 microliters and less than or equal to 50 microliters entries.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | {RangeP[0.5*Microliter, 50*Microliter]..}) | Automatic
Index Matches to: experiment samples
ReversePrimerVolume
The total volume of each reverse primer to add to the reaction.
Default Calculation: Automatically set to Null if ArrayCard is True, otherwise it is set to ForwardPrimerVolume (if specified) or 1 Microliter.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or list of one or more greater than or equal to 0.5 microliters and less than or equal to 50 microliters entries.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | {RangeP[0.5*Microliter, 50*Microliter]..}) | Automatic
Index Matches to: experiment samples
Probe
The short oligomer strand containing a reporter and quencher, which when separated during the synthesis reaction will result in fluorescence.
Default Calculation: Automatically set to the sample from the array card if ArrayCard is True, or Null otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Sample], Object[Sample]}] | _String) | {(ObjectP[{Model[Sample], Object[Sample]}] | _String)..}) | Automatic) | Null
Index Matches to: experiment samples
ProbeVolume
The volume of the probe to add to the reaction.
Default Calculation: Automatically set to Null if ArrayCard is True or Probe is Null, or 1 Microliter otherwise.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or list of one or more greater than or equal to 0.5 microliters and less than or equal to 50 microliters entries or Null.
Programmatic Pattern: ((RangeP[0.5*Microliter, 50*Microliter] | {RangeP[0.5*Microliter, 50*Microliter]..}) | Automatic) | Null
Index Matches to: experiment samples
EndogenousPrimerPair
The pair of short oligomer strands designed to amplify an endogenous control gene (such as a housekeeping gene) whose expression should not different between samples.
Pattern Description: {Forward Primer, Reverse Primer} or Null.
Programmatic Pattern: {ObjectP[{Model[Sample], Object[Sample]}] | _String, ObjectP[{Model[Sample], Object[Sample]}] | _String} | Null
Index Matches to: experiment samples
EndogenousForwardPrimerVolume
The total volume of each endogenous control forward primer to add to the reaction.
Default Calculation: Automatically resolves to match EndogenousReversePrimerVolume; otherwise, defaults to 1 Microliter.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: experiment samples
EndogenousReversePrimerVolume
The total volume of each endogenous control reverse primer to add to the reaction.
Default Calculation: Automatically resolves to match EndogenousForwardPrimerVolume; otherwise, defaults to 1 Microliter.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: experiment samples
EndogenousProbe
The short oligomer strand containing a reporter and quencher, which when separated during the synthesis reaction will result in fluorescence.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null
Index Matches to: experiment samples
EndogenousProbeVolume
The volume of the probe to add to the reaction.
Default Calculation: Automatically set to 1 Microliter if an endogenous probe is being used.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: experiment samples
NumberOfReplicates
Number of times each of the input samples should be analyzed using identical experimental parameters.
Pattern Description: Greater than or equal to 2 in increments of 1 or Null.
Programmatic Pattern: GreaterEqualP[2, 1] | Null
Post Experiment
AssayPlateStorageCondition
The conditions under which the thermocycled 384-well assay plate from this experiment should be stored after the protocol is completed. The assay plate is discarded by default.
Default Calculation: Automatically set to Disposal if using a 384-well plate, or Null otherwise.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
ArrayCardStorageCondition
The conditions under which the thermocycled array card from this experiment should be stored after the protocol is completed. The array card is discarded by default.
Default Calculation: Automatically set to Disposal if using an array card, or Null otherwise.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
MasterMixStorageCondition
The non-default condition under which the master mix used in this experiment should be stored after the protocol is completed. If left unset, the master mix will be stored according to its current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
StandardStorageCondition
The non-default conditions under which the standards of this experiment should be stored after the protocol is completed. If left unset, the standards will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standard
StandardForwardPrimerStorageCondition
The non-default conditions under which the forward primers for the standards of this experiment should be stored after the protocol is completed. If left unset, the forward primers for standards will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standard
StandardReversePrimerStorageCondition
The non-default conditions under which the reverse primers for the standards of this experiment should be stored after the protocol is completed. If left unset, the reverse primers for standards will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standard
StandardProbeStorageCondition
The non-default conditions under which the probes for the standards of this experiment should be stored after the protocol is completed. If left unset, the probes for standards will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standard
ForwardPrimerStorageCondition
The non-default conditions under which the forward primers of this experiment should be stored after the protocol is completed. If left unset, the forward primers will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
Index Matches to: experiment samples
ReversePrimerStorageCondition
The non-default conditions under which the reverse primers of this experiment should be stored after the protocol is completed. If left unset, the reverse primers will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
Index Matches to: experiment samples
ProbeStorageCondition
The non-default conditions under which the probes of this experiment should be stored after the protocol is completed. If left unset, the probes will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal)..}) | Null
Index Matches to: experiment samples
EndogenousForwardPrimerStorageCondition
The non-default conditions under which the endogenous forward primers of this experiment should be stored after the protocol is completed. If left unset, the endogenous forward primer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
EndogenousReversePrimerStorageCondition
The non-default conditions under which the endogenous reverse primers of this experiment should be stored after the protocol is completed. If left unset, the endogenous reverse primer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
EndogenousProbeStorageCondition
The non-default conditions under which the endogenous probes of this experiment should be stored after the protocol is completed. If left unset, the endogenous probe will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples
Reverse Transcription
ReverseTranscription
Indicates if one-step reverse transcription is performed in order to convert RNA input samples to cDNA.
Default Calculation: Automatically set to True if any reverse transcription related options are set, and False if none are set.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
ReverseTranscriptionTime
The length of time for which the reaction volume will held at ReverseTranscriptionTemperature in order to convert RNA to cDNA.
Default Calculation: Automatically set to 15 minutes if ReverseTranscription is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 2 hours or Null.
Programmatic Pattern: (RangeP[0*Second, 2*Hour] | Automatic) | Null
ReverseTranscriptionTemperature
The initial temperature at which the sample should be heated to in order to activate the reverse transcription enzymes in the master mix to convert RNA to cDNA.
Default Calculation: Automatically set to 48 degrees Celsius if ReverseTranscription is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[4*Celsius, 100*Celsius] | Automatic) | Null
ReverseTranscriptionRampRate
The rate at which the reaction volume is heated to bring the sample to the reverse transcription temperature.
Default Calculation: Automatically set to 1.6 Celsius/Second if ReverseTranscription is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 0.015 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second or Null.
Programmatic Pattern: (RangeP[0.015*(Celsius/Second), 3.4*(Celsius/Second)] | Automatic) | Null
Polymerase Activation
Activation
Indicates if polymerase hot start activation is performed. In order to reduce non-specific amplification, modern enzymes can be made room temperature stable by inhibiting their activity via thermolabile conjugates. Once an experiment is ready to be ran, this inhibition is removed by heating the reaction to ActivationTemperature.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
ActivationTime
The length of time for which the reaction volume is held at ActivationTemperature in order to a remove the thermolabile conjugates inhibiting enzyme activity.
Default Calculation: Automatically set to 1 minute if Activation is set to True.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 2 hours or Null.
Programmatic Pattern: (RangeP[0*Second, 2*Hour] | Automatic) | Null
ActivationTemperature
The temperature at which at which the thermolabile conjugates inhibiting enzyme activity are removed.
Default Calculation: Automatically set to 95 degrees Celsius if Activation is set to True.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[4*Celsius, 100*Celsius] | Automatic) | Null
ActivationRampRate
The rate at which the reaction is heated to bring the sample to the ActivationTemperature.
Default Calculation: Automatically set to 1.6 degrees Celsius per second if Activation is set to True.
Pattern Description: Greater than or equal to 0.015 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second or Null.
Programmatic Pattern: (RangeP[0.015*(Celsius/Second), 3.4*(Celsius/Second)] | Automatic) | Null
Denaturation
DenaturationTime
The length of time for which the reaction is held at DenaturationTemperature in order to separate any double stranded template DNA into single strands.
Default Value: 30 seconds
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 2 hours.
Programmatic Pattern: RangeP[0*Second, 2*Hour]
DenaturationTemperature
The temperature to which the sample is heated in order to disassociate double stranded template DNA into single strands.
Default Value: 95 degrees Celsius
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius.
Programmatic Pattern: RangeP[4*Celsius, 100*Celsius]
DenaturationRampRate
The rate at which the reaction is heated to bring the sample to DenaturationTemperature.
Default Value: 1.6 degrees Celsius per second
Pattern Description: Greater than or equal to 0.015 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second.
Programmatic Pattern: RangeP[0.015*(Celsius/Second), 3.4*(Celsius/Second)]
Primer Annealing
PrimerAnnealing
Indicates if annealing should be performed as a separate step instead of as part of extension. Lowering the temperature during annealing allows primers to bind to template DNA to serve as anchor points for DNA polymerization in the subsequent extension stage.
Default Calculation: Automatically set to True if any primer annealing related options are set, and False if none are set.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
PrimerAnnealingTime
The length of time for which the reaction is held at PrimerAnnealingTemperature in order to allow binding of primers to the template DNA.
Default Calculation: Automatically set to 30 second if PrimerAnnealing is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 2 hours or Null.
Programmatic Pattern: (RangeP[0*Second, 2*Hour] | Automatic) | Null
PrimerAnnealingTemperature
The temperature to which the sample is heated in order to allow primers to bind to the template strands.
Default Calculation: Automatically set to 60 Celsius if PrimerAnnealing is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[4*Celsius, 100*Celsius] | Automatic) | Null
PrimerAnnealingRampRate
The rate at which the reaction is heated to bring the sample to PrimerAnnealingTemperature.
Default Calculation: Automatically set to 1.6 Celsius/Second if PrimerAnnealing is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 0.015 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second or Null.
Programmatic Pattern: (RangeP[0.015*(Celsius/Second), 3.4*(Celsius/Second)] | Automatic) | Null
Strand Extension
ExtensionTime
The length of time for which the polymerase synthesizes a new DNA strand using the provided primer pairs and template DNA.
Default Value: 60 seconds
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 2 hours.
Programmatic Pattern: RangeP[0*Second, 2*Hour]
ExtensionTemperature
The temperature at which the sample is held to allow the polymerase to synthesis new DNA strand from the template DNA.
Default Value: 72 degrees Celsius
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius.
Programmatic Pattern: RangeP[4*Celsius, 100*Celsius]
ExtensionRampRate
The rate at which the reaction is heated to bring the sample to ExtensionTemperature.
Default Value: 1.6 degrees Celsius per second
Pattern Description: Greater than or equal to 0.015 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second.
Programmatic Pattern: RangeP[0.015*(Celsius/Second), 3.4*(Celsius/Second)]
NumberOfCycles
The number of times the samples will undergo repeated cycles of melting, annealing and extension.
Pattern Description: Greater than or equal to 1 and less than or equal to 60.
Programmatic Pattern: RangeP[1, 60]
Melting Curve
MeltingCurve
Indicates if melt curve should be performed at the end of the experiment. The fluorescence of the DuplexStainingDye is monitored while the sample is slowly heated, allowing detection of products with different melting temperatures.
Default Calculation: Automatically set to True for dye based assays and False to probe based assays.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
MeltingCurveStartTemperature
The temperature at which to start the melting curve.
Default Calculation: Automatically set to 60 Celsius if MeltingCurve is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[4*Celsius, 100*Celsius] | Automatic) | Null
MeltingCurveEndTemperature
The temperature at which to end the melting curve.
Default Calculation: Automatically set to 95 Celsius if MeltingCurve is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[4*Celsius, 100*Celsius] | Automatic) | Null
PreMeltingCurveRampRate
The rate at which the temperature is decreased in order to bring the samples to MeltingCurveStartTemperature.
Default Calculation: Automatically set to 1.6 Celsius/Second if MeltingCurve is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 0.002 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second or Null.
Programmatic Pattern: (RangeP[0.002*(Celsius/Second), 3.4*(Celsius/Second)] | Automatic) | Null
MeltingCurveRampRate
The rate at which the temperature is increased during in order to bring the samples from MeltingCurveStartTemperature to MeltingCurveEndTemperature.
Default Calculation: Automatically set to .015 Celsius/Second if MeltingCurve is set to True, and Null if it is set to False.
Pattern Description: Greater than or equal to 0.015 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second or Null.
Programmatic Pattern: (RangeP[0.015*(Celsius/Second), 3.4*(Celsius/Second)] | Automatic) | Null
MeltingCurveTime
The length of time it takes to change the temperature of the sample from MeltingCurveStartTemperature to MeltingCurveEndTemperature.
Default Calculation: Automatically set to a value based on MeltingCurveRampRate.
Pattern Description: Greater than or equal to 0.01 seconds and less than or equal to 24 hours or Null.
Programmatic Pattern: (RangeP[0.01*Second, 24*Hour] | Automatic) | Null
Dye Staining
DuplexStainingDye
The fluorescent dye present in the reaction which intercalates between the base pairs of a double stranded DNA.
Default Calculation: Automatically set to the duplex stain dye part of the MasterMix for dye based assay or Null if one isn't present.
Pattern Description: An object of type or subtype Object[Sample], Model[Sample], or Model[Molecule] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample], Model[Molecule]}] | _String) | Automatic) | Null
DuplexStainingDyeVolume
The volume of the fluorescent dye to add to the reaction.
Default Calculation: Automatically set to Null if the dye is already part of the master mix or 1 Microliter if it is being added separately.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 200*Microliter] | Automatic) | Null
ReferenceDye
The fluorescent dye present in the reaction which does not takes part of the reaction and is used for signal normalization.
Default Calculation: Automatically set to the passive dye present in the master mix.
Pattern Description: An object of type or subtype Object[Sample], Model[Sample], or Model[Molecule] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample], Model[Molecule]}] | _String) | Automatic) | Null
ReferenceDyeVolume
The volume of the fluorescent reference dye to add to the reaction.
Default Calculation: Automatically set to Null when using a pre-made master mix.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Detection
DuplexStainingDyeExcitationWavelength
The wavelength of light used to excite the fluorescent reporter dye.
Default Calculation: Automatically set to the wavelength excitation wavelength of the DuplexStainingDye
Pattern Description: 470 nanometers, 520 nanometers, 550 nanometers, 580 nanometers, 640 nanometers, or 662 nanometers or Null.
Programmatic Pattern: (qPCRExcitationWavelengthP | Automatic) | Null
DuplexStainingDyeEmissionWavelength
The wavelength of light emitted by the fluorescent reporter dye once it has been excited and read by the instrument.
Default Calculation: Automatically set to the emission wavelength of the DuplexStainingDye
Pattern Description: 520 nanometers, 558 nanometers, 586 nanometers, 623 nanometers, 682 nanometers, or 711 nanometers or Null.
Programmatic Pattern: (qPCREmissionWavelengthP | Automatic) | Null
ReferenceDyeExcitationWavelength
The wavelength of light used to excite the fluorescent reference dye.
Default Calculation: Automatically set to the wavelength excitation wavelength of the ReferenceDye
Pattern Description: 470 nanometers, 520 nanometers, 550 nanometers, 580 nanometers, 640 nanometers, or 662 nanometers or Null.
Programmatic Pattern: (qPCRExcitationWavelengthP | Automatic) | Null
ReferenceDyeEmissionWavelength
The wavelength of light emitted by the fluorescent reference dye once it has been excited and read by the instrument.
Default Calculation: Automatically set to the emission wavelength of the ReferenceDye
Pattern Description: 520 nanometers, 558 nanometers, 586 nanometers, 623 nanometers, 682 nanometers, or 711 nanometers or Null.
Programmatic Pattern: (qPCREmissionWavelengthP | Automatic) | Null
StandardProbeFluorophore
The fluorescent molecule conjugated to the standard probe that allows for detection of amplification.
Pattern Description: An object of type or subtype Model[Molecule] or Null.
Programmatic Pattern: (ObjectP[Model[Molecule]] | Automatic) | Null
Index Matches to: Standard
StandardProbeExcitationWavelength
The wavelength of light used to excite the reporter component of the standard probe.
Default Calculation: Automatically set to a wavelength appropriate to the fluorophore attached to the standard probe.
Pattern Description: 470 nanometers, 520 nanometers, 550 nanometers, 580 nanometers, 640 nanometers, or 662 nanometers or Null.
Programmatic Pattern: (qPCRExcitationWavelengthP | Automatic) | Null
Index Matches to: Standard
StandardProbeEmissionWavelength
The wavelength of light emitted by the reporter once it has been excited and read by the instrument.
Default Calculation: Automatically set to a wavelength appropriate to the fluorophore attached to the standard probe.
Pattern Description: 520 nanometers, 558 nanometers, 586 nanometers, 623 nanometers, 682 nanometers, or 711 nanometers or Null.
Programmatic Pattern: (qPCREmissionWavelengthP | Automatic) | Null
Index Matches to: Standard
ProbeFluorophore
The fluorescent molecule conjugated to the probe that allows for detection of amplification.
Default Calculation: Automatically set from the array card model if ArrayCard is True, or Null otherwise.
Pattern Description: An object of type or subtype Model[Molecule] or list of one or more an object of type or subtype Model[Molecule] entries or Null.
Programmatic Pattern: ((ObjectP[Model[Molecule]] | {ObjectP[Model[Molecule]]..}) | Automatic) | Null
Index Matches to: experiment samples
ProbeExcitationWavelength
The wavelength of light used to excite the reporter component of the probe.
Default Calculation: Automatically set to a wavelength appropriate to the fluorophore attached to the probe.
Pattern Description: 470 nanometers, 520 nanometers, 550 nanometers, 580 nanometers, 640 nanometers, or 662 nanometers or list of one or more 470 nanometers, 520 nanometers, 550 nanometers, 580 nanometers, 640 nanometers, or 662 nanometers entries or Null.
Programmatic Pattern: ((qPCRExcitationWavelengthP | {qPCRExcitationWavelengthP..}) | Automatic) | Null
Index Matches to: experiment samples
ProbeEmissionWavelength
The wavelength of light emitted by the reporter once it has been excited and read by the instrument.
Default Calculation: Automatically set to a wavelength appropriate to the fluorophore attached to the probe.
Pattern Description: 520 nanometers, 558 nanometers, 586 nanometers, 623 nanometers, 682 nanometers, or 711 nanometers or list of one or more 520 nanometers, 558 nanometers, 586 nanometers, 623 nanometers, 682 nanometers, or 711 nanometers entries or Null.
Programmatic Pattern: ((qPCREmissionWavelengthP | {qPCREmissionWavelengthP..}) | Automatic) | Null
Index Matches to: experiment samples
EndogenousProbeFluorophore
The fluorescent molecule conjugated to the endogenous probe that allows for detection of amplification.
Pattern Description: An object of type or subtype Model[Molecule] or Null.
Programmatic Pattern: (ObjectP[Model[Molecule]] | Automatic) | Null
Index Matches to: experiment samples
EndogenousProbeExcitationWavelength
The wavelength of light used to excite the reporter component of the endogenous probe.
Default Calculation: Automatically set to a wavelength appropriate to the fluorophore attached to the endogenous probe.
Pattern Description: 470 nanometers, 520 nanometers, 550 nanometers, 580 nanometers, 640 nanometers, or 662 nanometers or Null.
Programmatic Pattern: (qPCRExcitationWavelengthP | Automatic) | Null
Index Matches to: experiment samples
EndogenousProbeEmissionWavelength
The wavelength of light emitted by the reporter once it has been excited and read by the instrument.
Default Calculation: Automatically set to a wavelength appropriate to the fluorophore attached to the endogenous probe.
Pattern Description: 520 nanometers, 558 nanometers, 586 nanometers, 623 nanometers, 682 nanometers, or 711 nanometers or Null.
Programmatic Pattern: (qPCREmissionWavelengthP | Automatic) | Null
Index Matches to: experiment samples
Standard Curve
Standard
The sample that is serially diluted in order to construct a standard curve.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null
Index Matches to: Standard
StandardPrimerPair
The primer pair used to amplify a target sequence in Standard.
Pattern Description: {Forward Primer, Reverse Primer} or {Null, Null} or Null.
Programmatic Pattern: ({ObjectP[{Model[Sample], Object[Sample]}] | _String, ObjectP[{Model[Sample], Object[Sample]}] | _String} | {Null, Null}) | Null
Index Matches to: Standard
SerialDilutionFactor
The factor by which the standard should be serially diluted with Buffer.
Default Calculation: Resolved automatically to 2, meaning that each standard serial dilution will be twofold.
Pattern Description: Greater than or equal to 1 and less than or equal to 100 or Null.
Programmatic Pattern: (RangeP[1, 100] | Automatic) | Null
Index Matches to: Standard
NumberOfDilutions
The number of times the standard should serially diluted, including the original (undiluted) standard sample.
Default Calculation: Resolved automatically to 8, meaning the standard curve will consist of the undiluted standard sample followed by 7 serial dilutions by SerialDilutionFactor.
Pattern Description: Greater than or equal to 1 and less than or equal to 12 or Null.
Programmatic Pattern: (RangeP[1, 12] | Automatic) | Null
Index Matches to: Standard
NumberOfStandardReplicates
The number of times the standard should be replicated per each unique set of primer target pairs.
Default Calculation: Resolved automatically to 1.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 or Null.
Programmatic Pattern: (RangeP[1, 10] | Automatic) | Null
Index Matches to: Standard
StandardForwardPrimerVolume
The total volume of each forward primer to add to the standard reaction.
Default Calculation: Automatically resolves to match StandardReversePrimerVolume; otherwise, defaults to 1 Microliter.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardReversePrimerVolume
The total volume of each reverse primer to add to the standard reaction.
Default Calculation: Automatically resolves to match StandardForwardPrimerVolume; otherwise, defaults to 1 Microliter.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardProbe
The short oligomer strand containing a reporter and quencher, which when separated during the synthesis reaction will result in fluorescence.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null
Index Matches to: Standard
StandardProbeVolume
The volume of the standard probe to add to the reaction.
Default Calculation: Automatically set to 1 Microliter if a Standard Probe is being used.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: Standard
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and when the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise set to Model[Container, Vessel, "2mL Tube"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[Model[Container]] | Automatic) | Null
Index Matches to: experiment samples
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
Index Matches to: experiment samples
Sample Handling
MoatSize
Indicates the number of concentric perimeters of wells which should be should be filled with MoatBuffer in order to decrease evaporation from the assay samples during the run.
Pattern Description: Greater than 0 in increments of 1 or Null.
Programmatic Pattern: (GreaterP[0, 1] | Automatic) | Null
MoatVolume
Indicates the volume which should be added to each moat well.
Default Calculation: Automatically set to the RecommendedFillVolume of the assay plate if informed, or 75% of the MaxVolume of the assay plate if not, if any other moat options are specified.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
MoatBuffer
Indicates the buffer which should be used to fill each moat well.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null