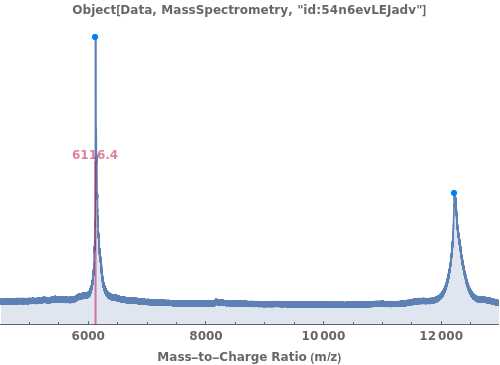
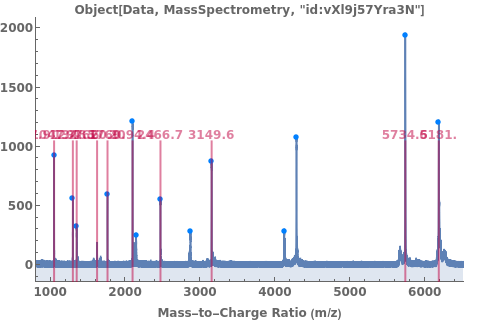
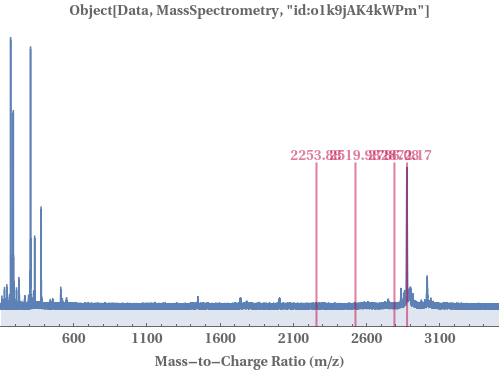
PlotMassSpectrometry
PlotMassSpectrometry[massSpectrometryData]⟹plot
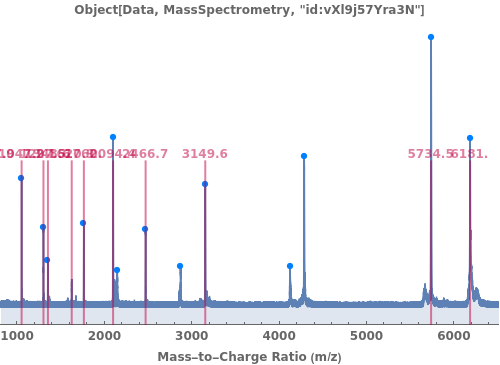
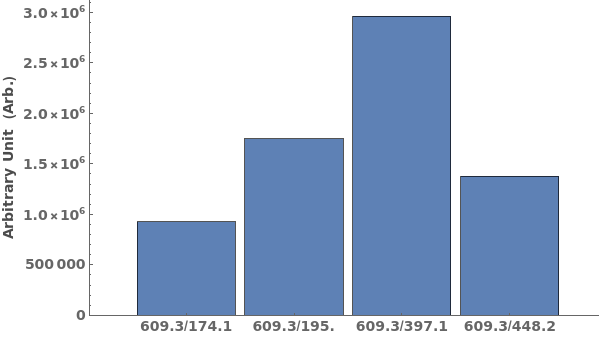
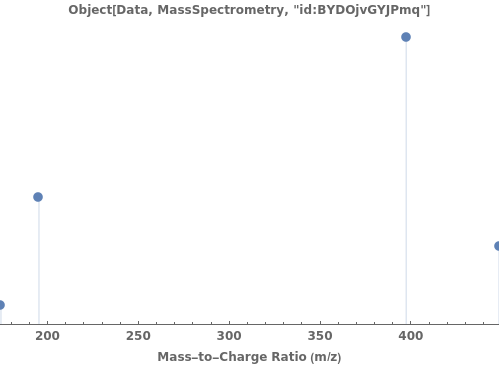
provides a graphical plot the provided mass spectra.
PlotMassSpectrometry[spectra]⟹plot
provides a graphical plot the provided mass spectra.
Details
Input
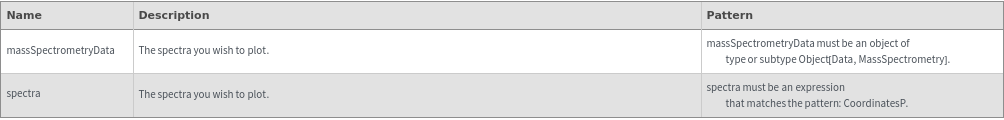
Output

Data Specifications Options
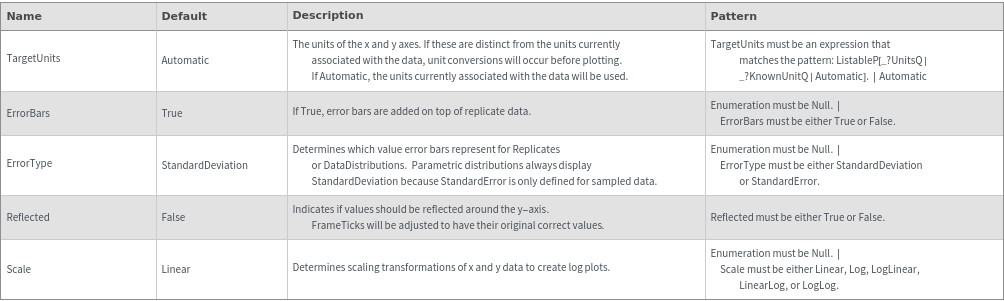
Fractions Options

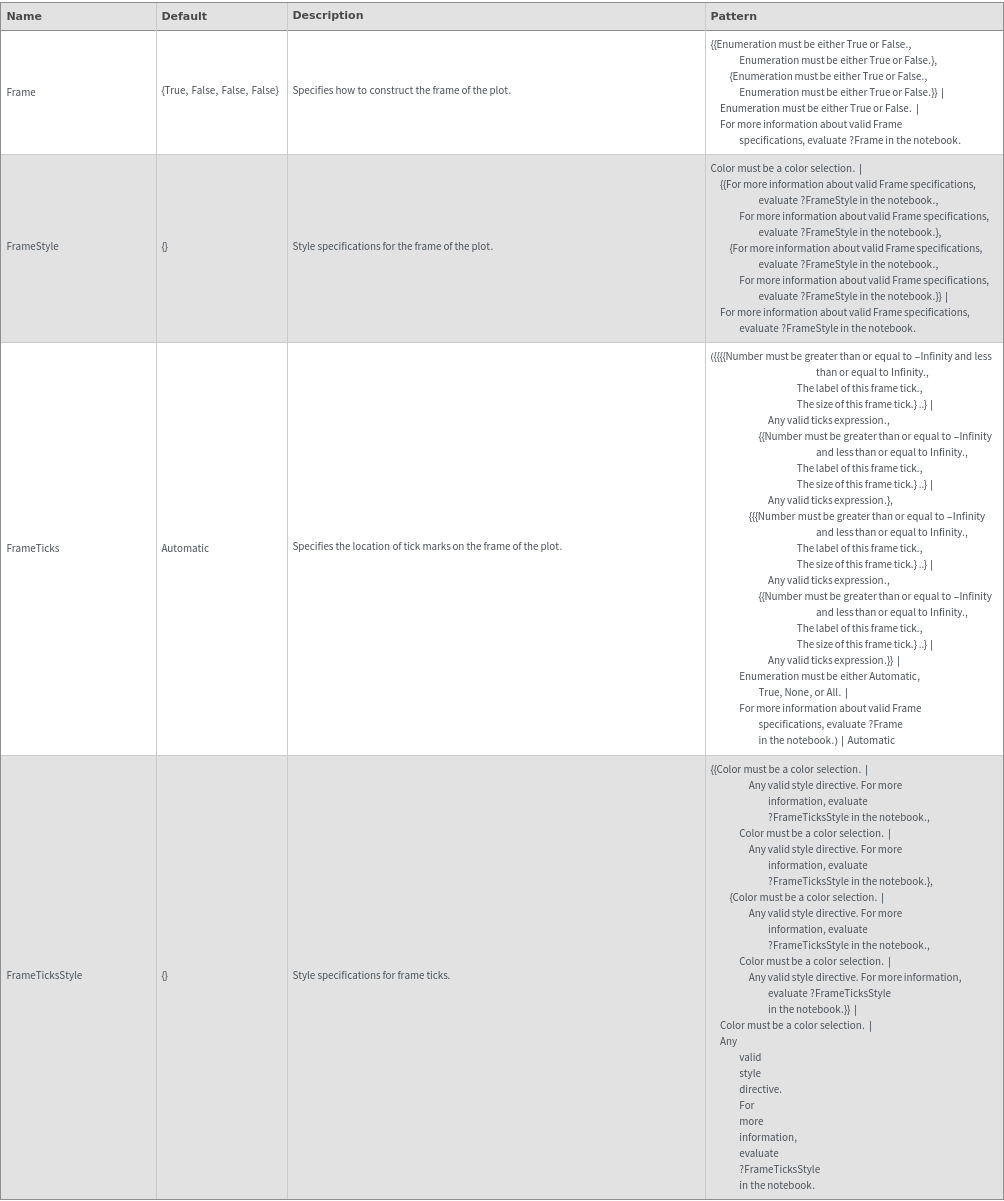
Frame Options
Grid Options
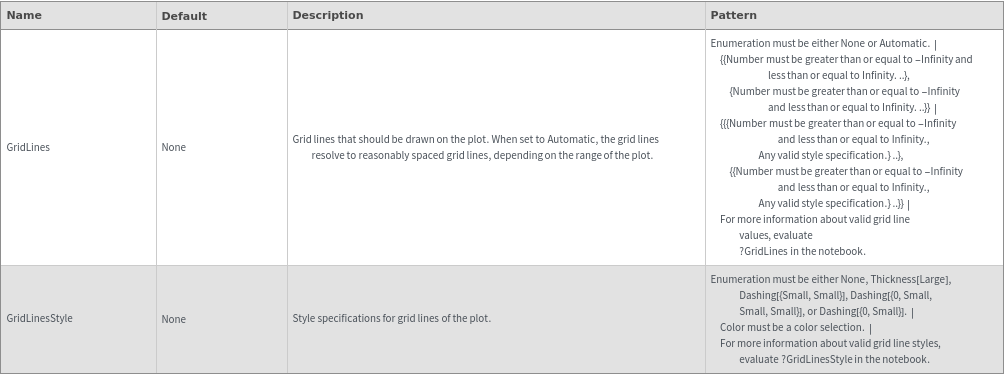
Image Format Options
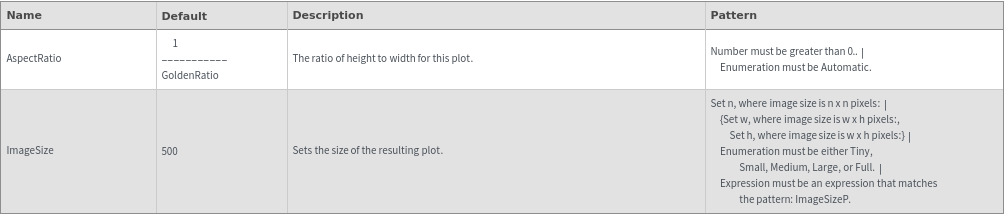
Ladder Options
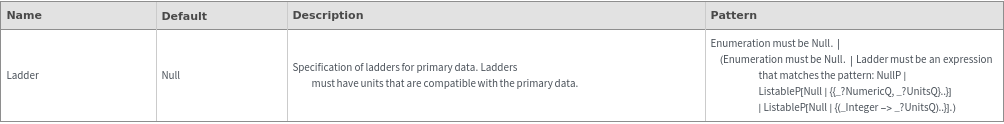
Legend Options
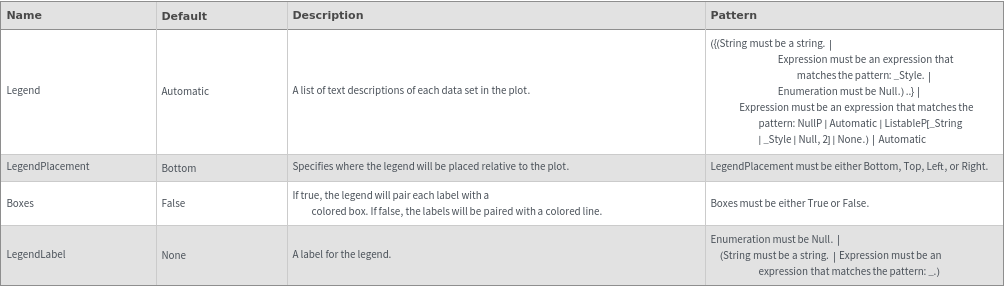
MolecularWeightEpilog Options
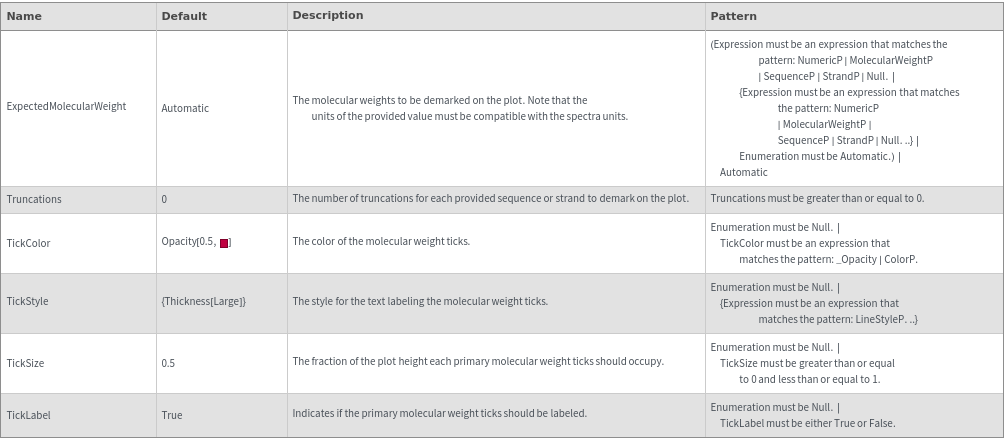
Peaks Options
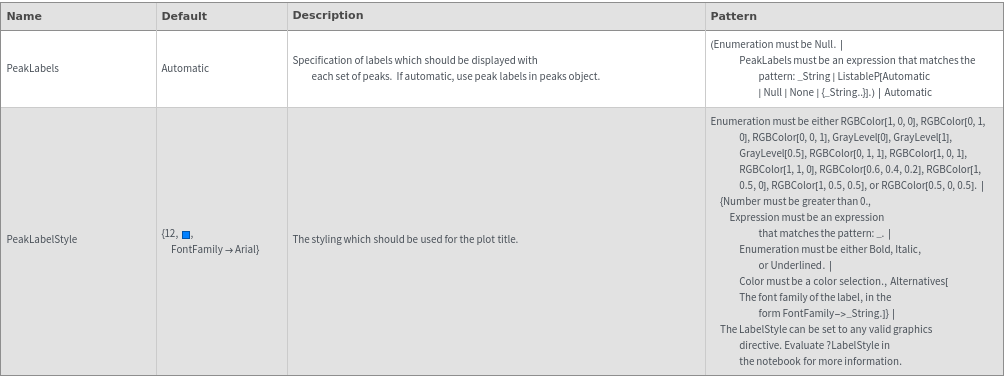
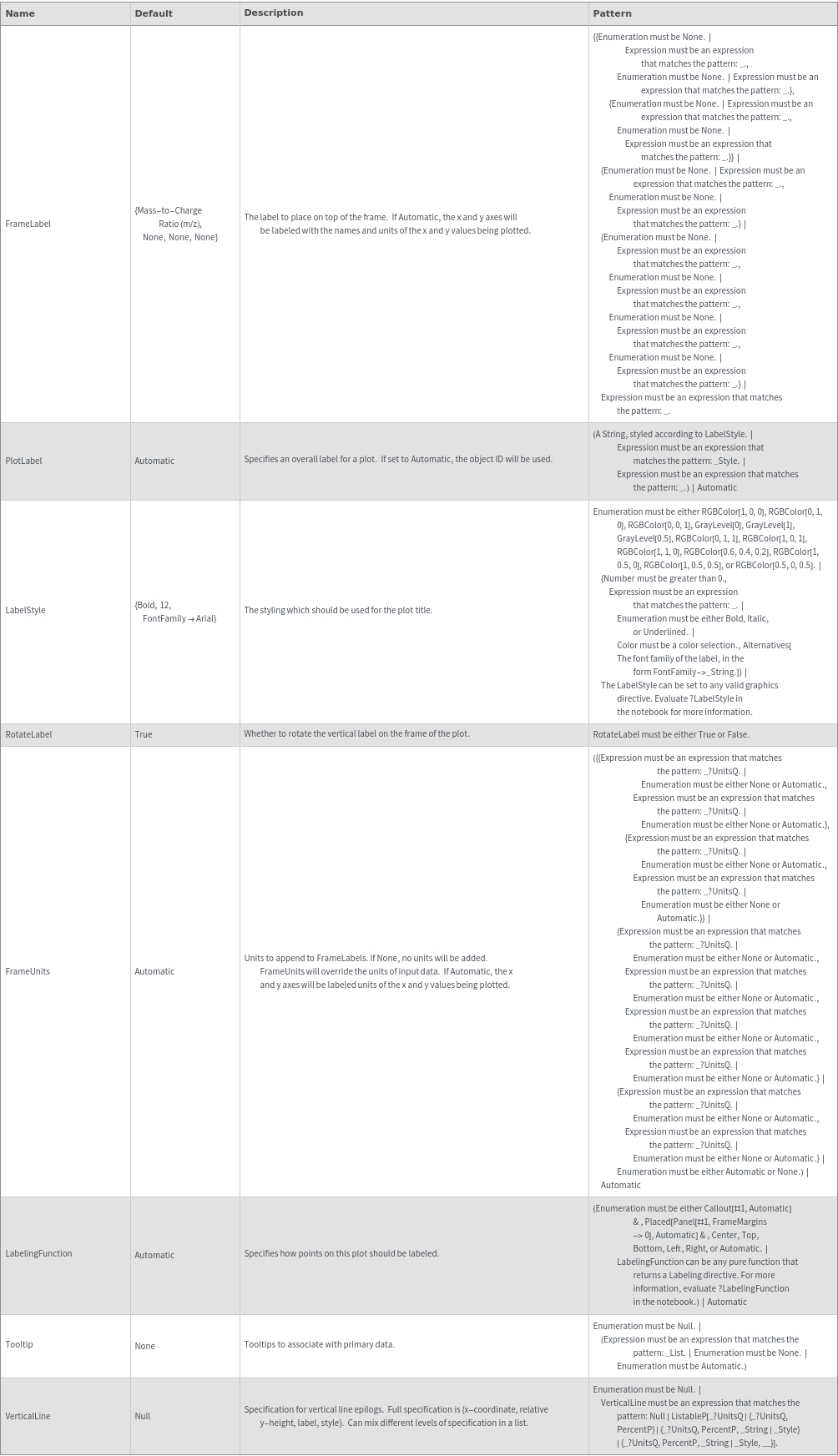
Plot Labeling Options
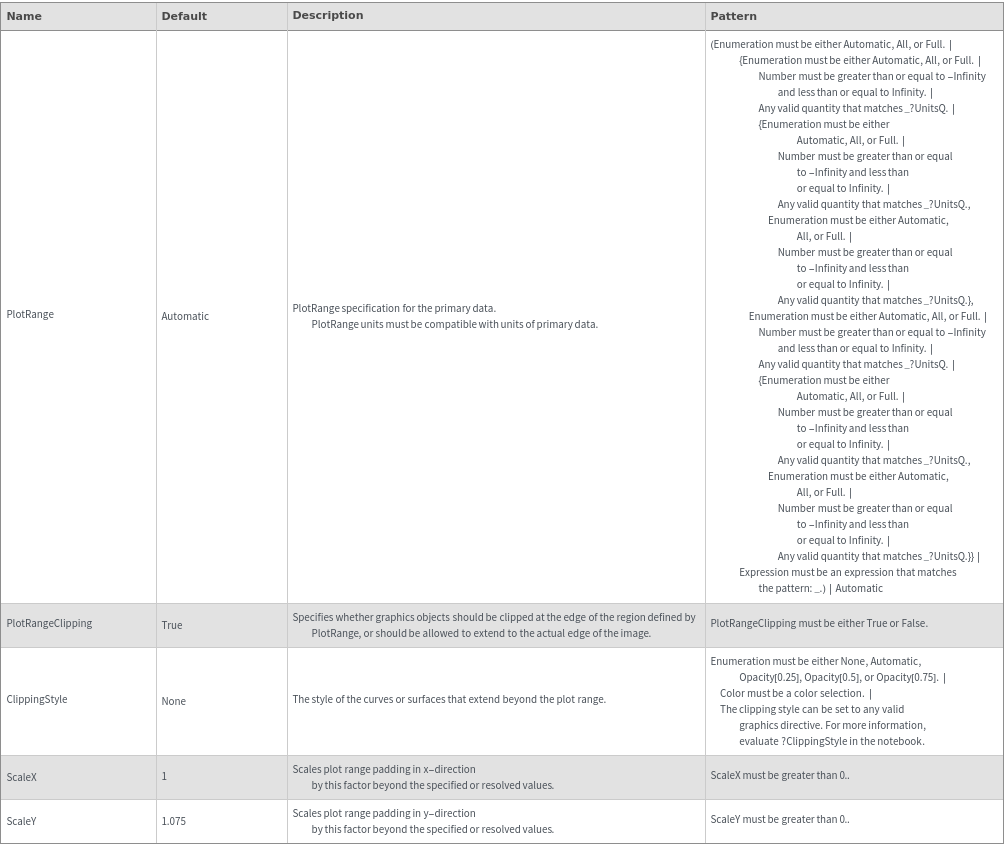
Plot Range Options
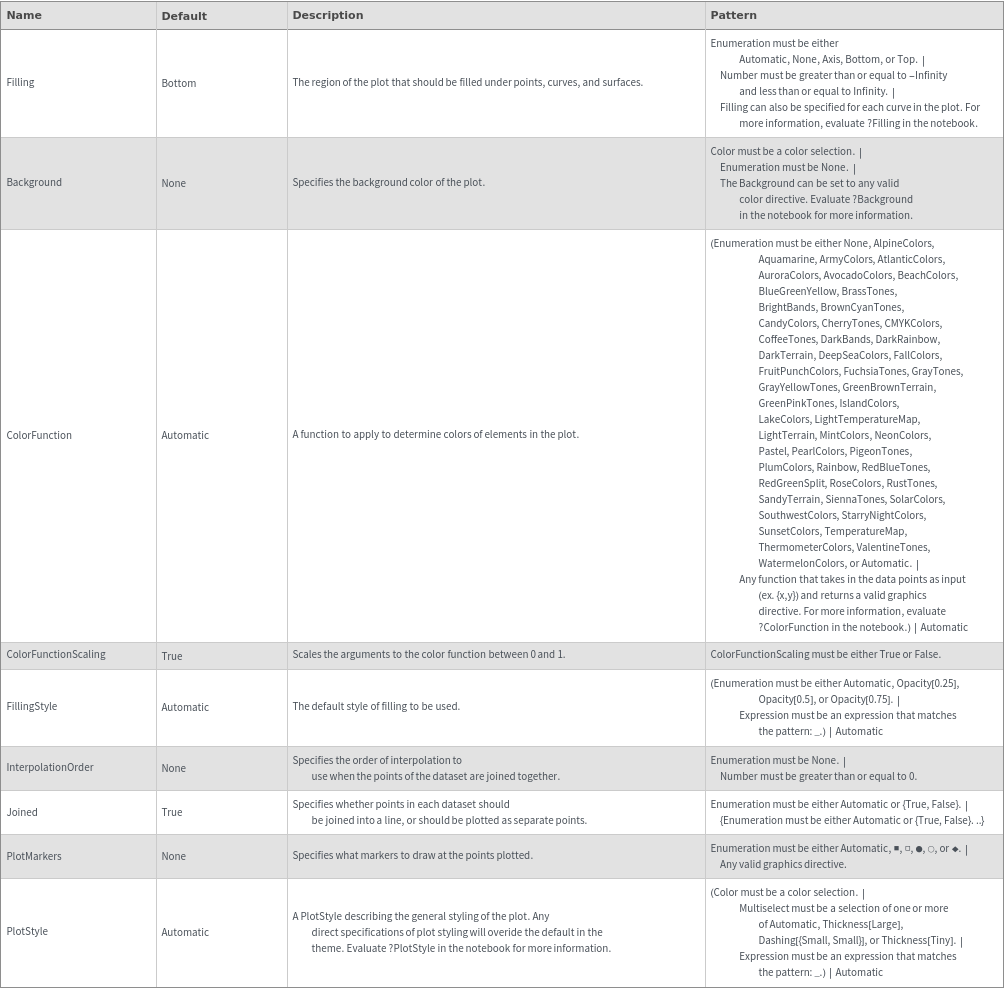
Plot Style Options
Secondary Data Options
General Options