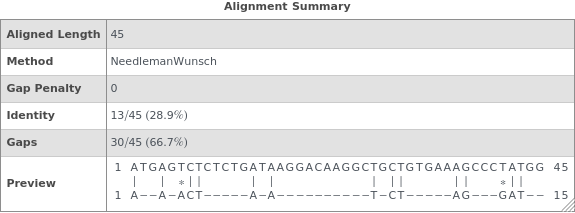
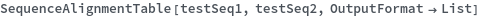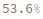SequenceAlignmentTable
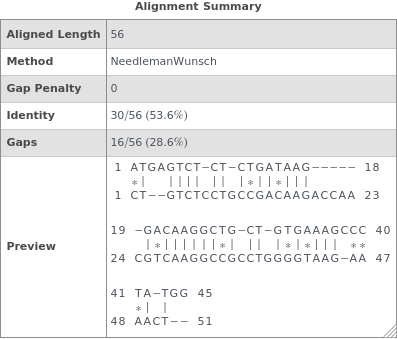
SequenceAlignmentTable[sequence1,sequence2]⟹summary
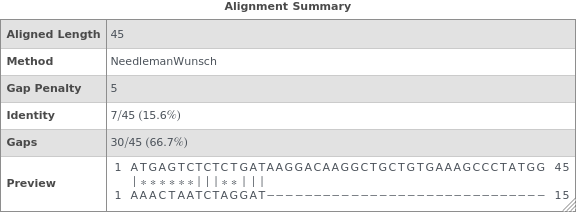
returns a table showing sequence similarity, identity, gaps, and a visual preview of the alignment betwen sequence1 and sequence2.
Examples
Basic Examples (4)
Options (11)
GapPenalty (2)
By default, the GapPenalty is zero and a global alignment is used. If one sequence is much shorter than the other, this shorter sequence can be substantially fragmented in the alignment:

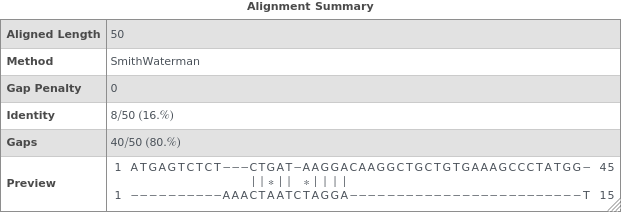
Increasing the GapPenalty reduces the number of discontinuous gaps in the alignment at the expense of decreasing the number of matches: