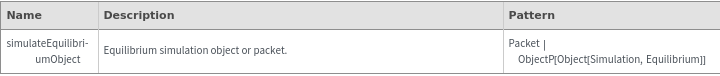
SimulateEquilibrium
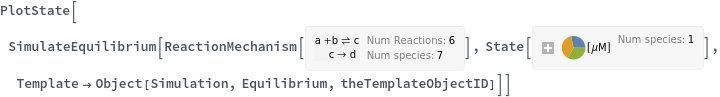
SimulateEquilibrium[reactionModel, initialCondition]⟹simulateEquilibriumObject
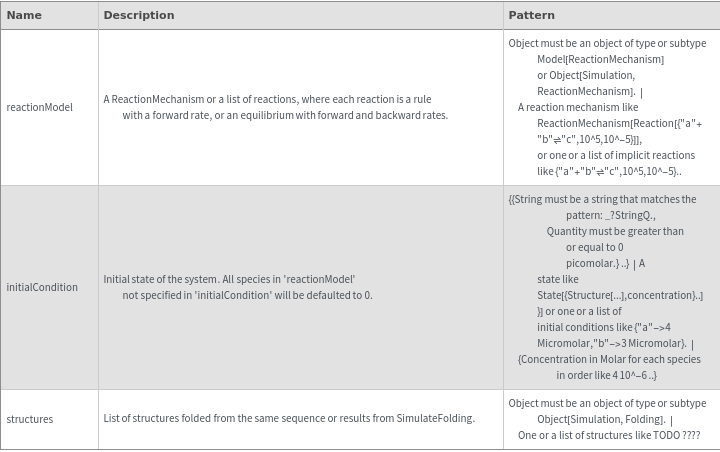
computes the equilibrium of the ReactionMechanism reactionModel starting from initialCondition condition initialCondition.
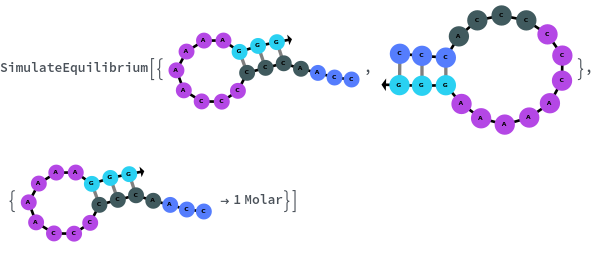
SimulateEquilibrium[structures, initialCondition]⟹simulateEquilibriumObject
constructs a ReactionMechanism from structures under two-state assumption then calculate equilibrium state.
Details
- Given a set of reactions and their associated rate constants, and an initial concentration of each species, SimulateEquilibrium solves the system of equations to determine concentrations of the species after all of the reactions have proceeded to equilibrium.
- When a list of nucleic acid structures is provided as input, SimulateEquilibrium assumes that the ReactionMechanism is two-state, i.e. each structure is generated only from fully unfolded structure, and there's no cross talk between the structures or any intermediate states.
- When input is a list of nucleic acid structures or a reactionModel without explicit reaction rates, assumes binding rate constant for binding at 5*10^5 (Molar*Second)^-1 and calculates the melting rate based on the detailed balance (Keq = kf / kb) along with the nearest-neighbor thermodynamics simulation of equilibrium constant for binding (see SimulateEquilibriumConstant).
-
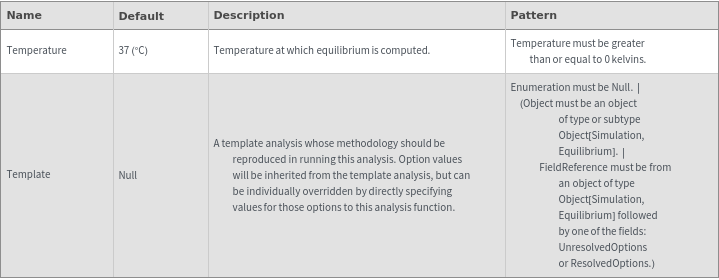
NegativeConcentration The equilibrium state contains speices with substantial negative concentrations.
Input
Output
General Options
Messages
Examples
Basic Examples (3)
Additional Examples (6)
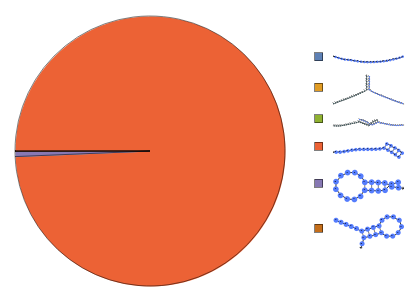
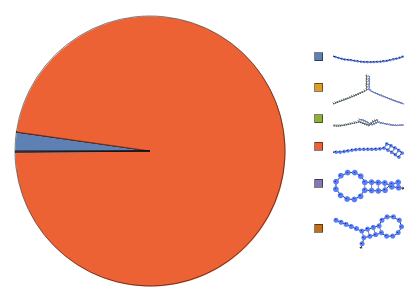
Input a ReactionMechanism model and a list of initial conditions:

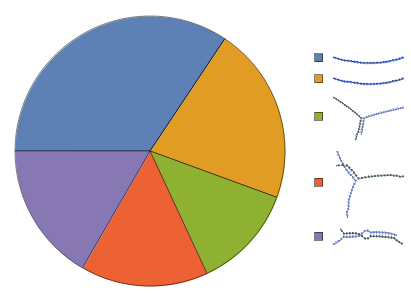
Construct a ReactionMechanism from folding results under two-state assumption then calculate equilibrium:

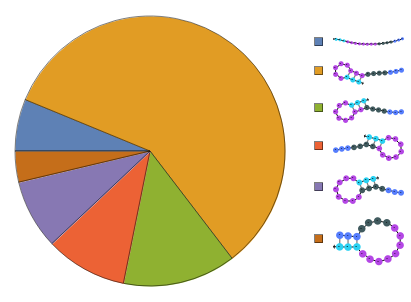
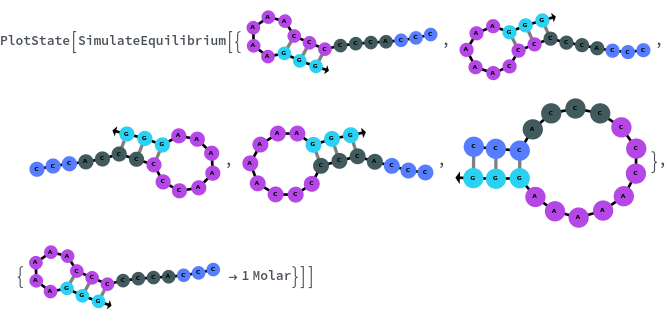
Construct a ReactionMechanism on a list of structures folded from the same sequence under two-state assumption then calculate equilibrium:

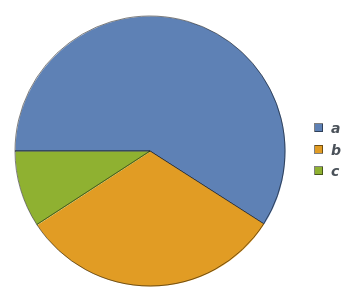
If input is a single reaction, it will be treated as a ReactionMechanism with only one reaction:

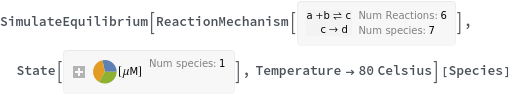
Return all species in the reaction system:
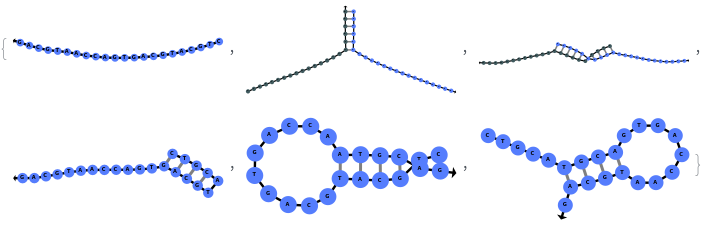
Return both initial and equilibrium states: