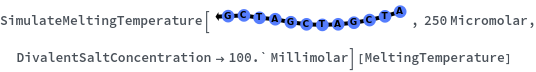
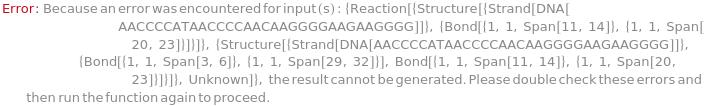
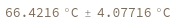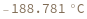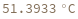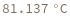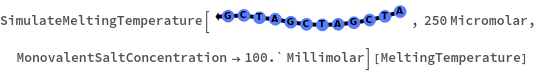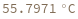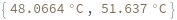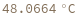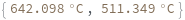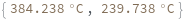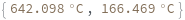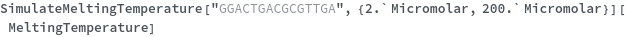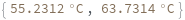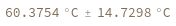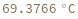SimulateMeltingTemperature
SimulateMeltingTemperature[reaction, concentration]⟹meltingTemperatureObject
computes the melting temperature of the given reaction between two nucleic acid oligomers at the specified concentration with traditional Nearest Neighbor thermodynamic analysis.
SimulateMeltingTemperature[reactantAplusB, concentration]⟹meltingTemperatureObject
finds the product of reaction from 'reactantA' + 'reactantB', then computes the melting temperature.
SimulateMeltingTemperature[reactantEquilibriumProduct, concentration]⟹meltingTemperatureObject
infers the type of reaction from the given 'reactant' ⇌ 'product' state and computes the melting temperature for that reaction.
SimulateMeltingTemperature[reactionMechanism, concentration]⟹meltingTemperatureObject
computes the melting temperature from the reaction in the given mechanism.
SimulateMeltingTemperature[oligomer, concentration]⟹meltingTemperatureObject
considers the hybridization reaction between the given oligomer and its reverse complement.
SimulateMeltingTemperature[structure, concentration]⟹meltingTemperatureObject
considers the melting reaction whereby all of the bonds in the given structure are melted.
SimulateMeltingTemperature[enthalpy, entropy, concentration]⟹meltingTemperatureObject
computes the melting temperature from the given enthalpy and entropy of a reaction.
Details
- Melting temperature is defined as the temperature at which half of the strands are in the double-helical state and half are in the random-coil state.
- In first order folding or melting case, melting temperature is calculated from Tm = ΔH/ΔS.
- In second order paring or melting case, melting temperature is calculated from Tm = ΔH/(ΔS-R ln[Ct]). For bimolecular self-complementary cases (A + A ⇌ A2), Ct = C[A] (C[A] represents the concentration of A). For bimolecular nonself-complimentary cases (A + B ⇌ AB), if C[A] = C[B], Ct = (C[A] + C[B]) / 4, if C[A] > C[B], Ct = C[A] - C[B]/2, and if C[A] < C[B], Ct = C[B] - C[A]/2. See reference: Object[Report,Literature,"id:o1k9jAKpjE8a"]: John SantaLucia. "A Unified View of Polymer,Dumbbell,and Oligonucleotide DNA Nearest-Neighbor Thermodynamics." Proceedings of the National Academy of Sciences of the United States of America 95.4 (1998):1460–1465.
- DNA Nearest Neighbor parameters from Object[Report, Literature, "id:kEJ9mqa1Jr7P"]: Allawi, Hatim T., and John SantaLucia. "Thermodynamics and NMR of internal GT mismatches in DNA." Biochemistry 36.34 (1997): 10581-10594.
- RNA Nearest Neighbor parameters from Object[Report, Literature, "id:M8n3rxYAnNkm"]: Xia, Tianbing, et al. "Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs." Biochemistry 37.42 (1998): 14719-14735.
- If given a nucleic acid sequence, strand, or sequence length, this function assumes a two-state binding between the provided sequence and a perfect reverse complement.
- Given a structure, considers only the bonded regions of the structure.
- Supported polymer types are DNA and RNA.
- Untyped sequences or lengths default to DNA if there is ambiguity.
- Enthalpy is independent of salt concentration, while entropy values for a given salt concentration. MonovalentSaltConcentration and DivalentSaltConcentration can be used to specify the concentration of monovalent salt (Na+, K+) and divalent salt (Mg2+) respectively. The entropy correction term is calculated as: 0.368*(Sequence Length - 1)*ln[(Na+) + 140*(Mg2+)] from Object[Report,Literature, "id:eGakld09nLXo"]: von Ahsen, et al. "Application of a Thermodynamic Nearest-Neighbor Model to Estimate Nucleic Acid Stability and Optimize Probe Design:Prediction of Melting Points of Multiple Mutations of Apolipoprotein B-3500 and Factor V with a Hybridization Probe Genotyping Assay on the LightCycler" Clinical Chemistry 45.12 (1999) 2094-2101.
- Assumes a second order reaction if enthalpy and entropy are provided.
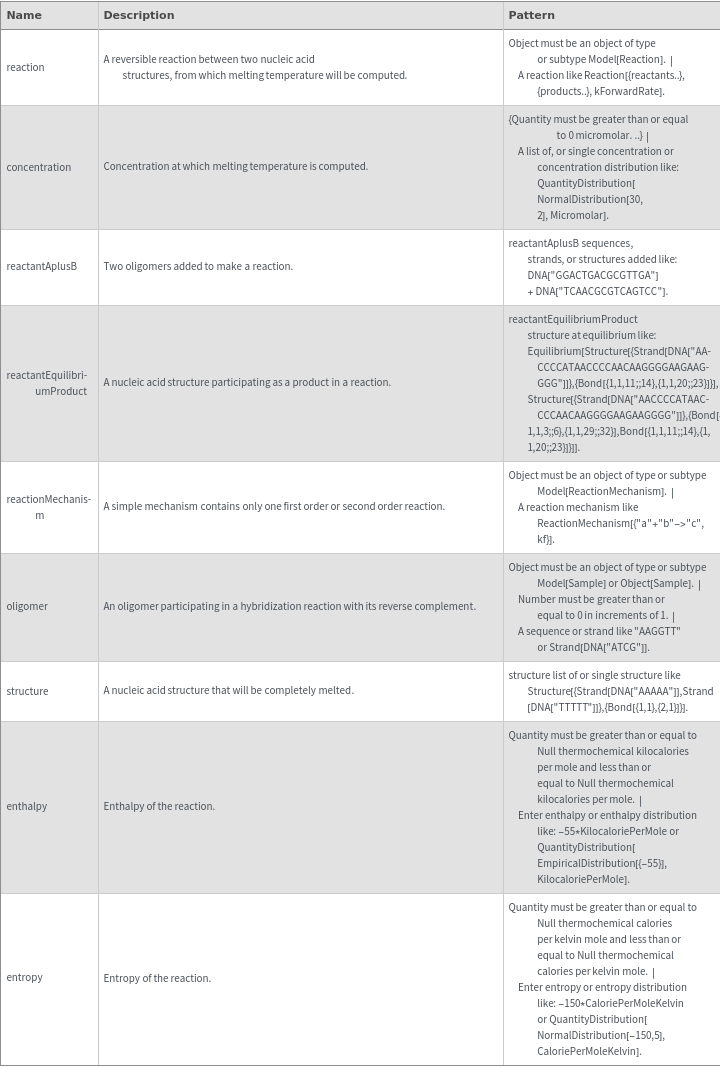
Input
Output

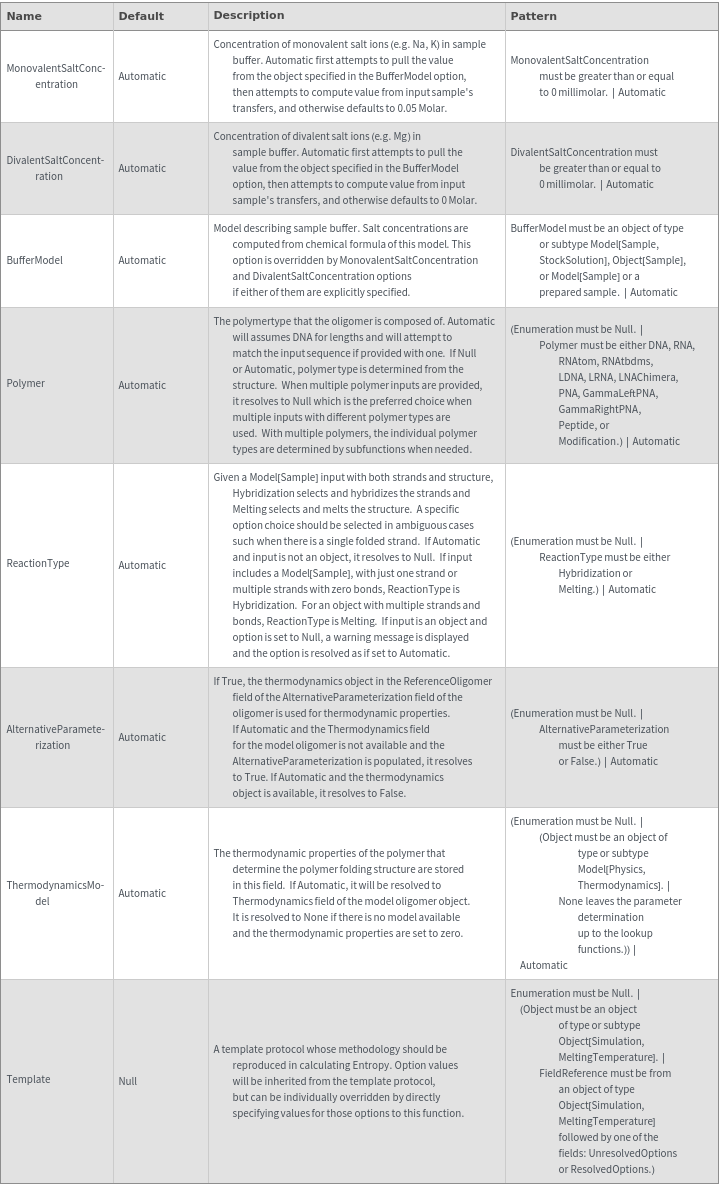
General Options
Examples
Basic Examples (5)
Compute the melting temperature of a hybridization reaction between given sequence and its reverse complement:

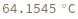
Find the product of DNA['GGACTGACGCGTTGA']+DNA['TCAACGCGTCAGTCC'], then compute the melting temperature:

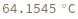
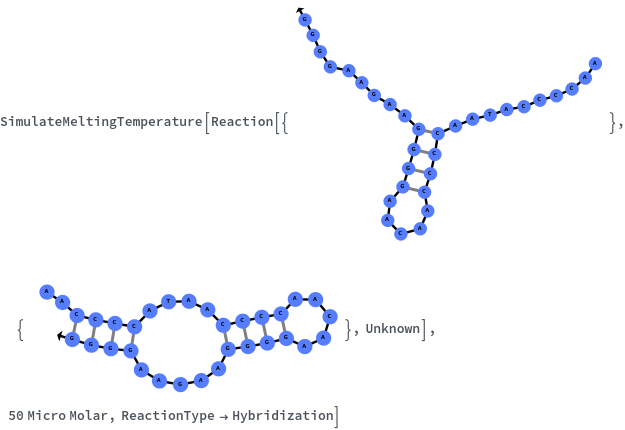
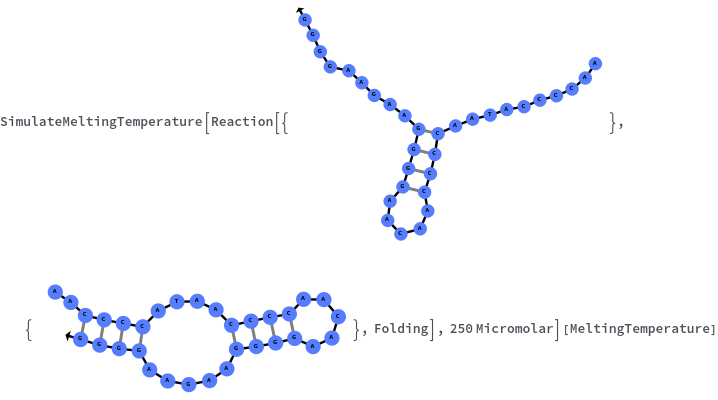
Specify reaction from one structure to another:
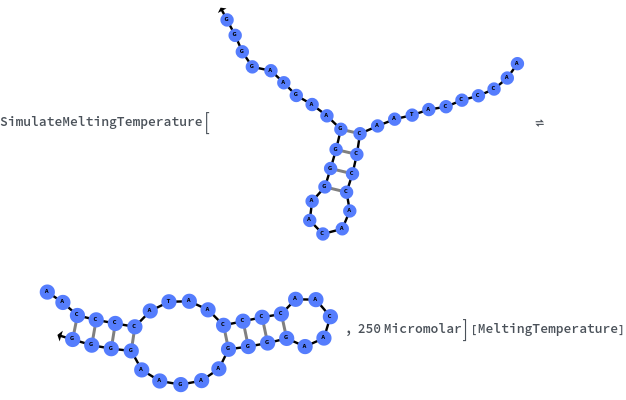
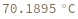
Specify reaction from one structure to another:
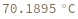
Compute the melting temperature of a bimolecular reaction from its enthalpy and entropy:

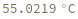
Additional Examples (17)
Input concentration as a distribution:

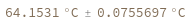
Two reactants have different concentrations:

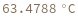
Two reactants have different concentrations and one of them is specified as a distribution:

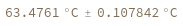
Two reactants have different concentrations as distributions:

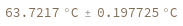
Input enthalpy, entropy and concentration as distributions:

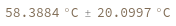
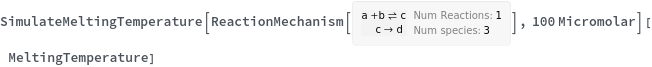
Compute the melting temperature from a simple ReactionMechanism contains only one reaction:
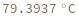
Pull strand from given sample:

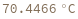

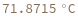

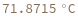
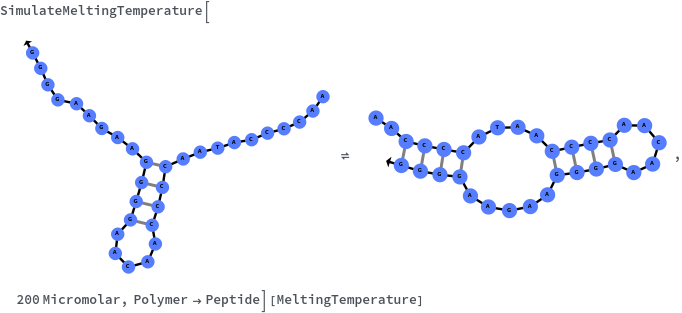
Given structure, computes melting temperature of all bonded regions:
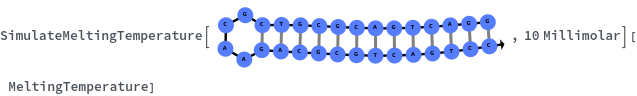
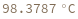
Compute the distribution of melting temperature of all 15-mer hybridization reactions with their reverse complements:

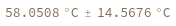
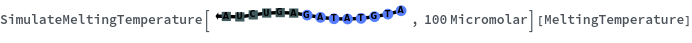
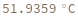

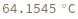

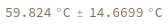
Structure with no bonds returns 0K:
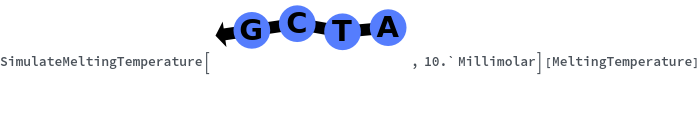
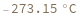
Can handle degenerate sequence:

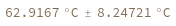
Return melting temperature distribution instead of mean: