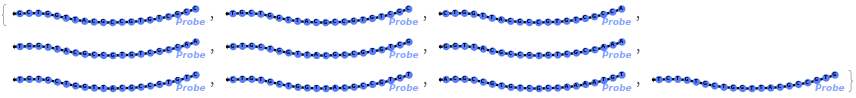
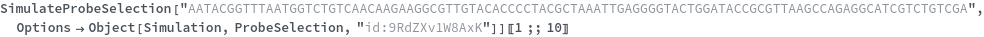SimulateProbeSelection
SimulateProbeSelection[target]⟹probeStrand
given a target sequence, simulates which probe sequence would most faithfully bind to the target at any site along the target sequence by performing kinetic simulations taking into account any folding or self-pairing of the probe or target sequences.
SimulateProbeSelection[target, sites]⟹probeStrand
given a target sequence, and sites on the target to bind to, simulates which probe sequence would most faithfully bind to the target's sites along the target sequence by performing kinetic simulations taking into account any folding, self-pairing, or miss-hybridization of the probe to the target at just the sites specified.
SimulateProbeSelection[transcript]⟹probeStrand
a ribonuleic acid (RNA) transcript can be directly applied into the function to simulate which probe sequence would most faithfully bind to the target.
Details
- For each site being tested a mechanism is generated (via SimulateReactivity) to model the interactions between the probe and target. The types of interactions considered in the mechanism are determined by the options ProbeTarget, ProbeFolding, ProbeHybridization, TargetFolding, and TargetHybridization.
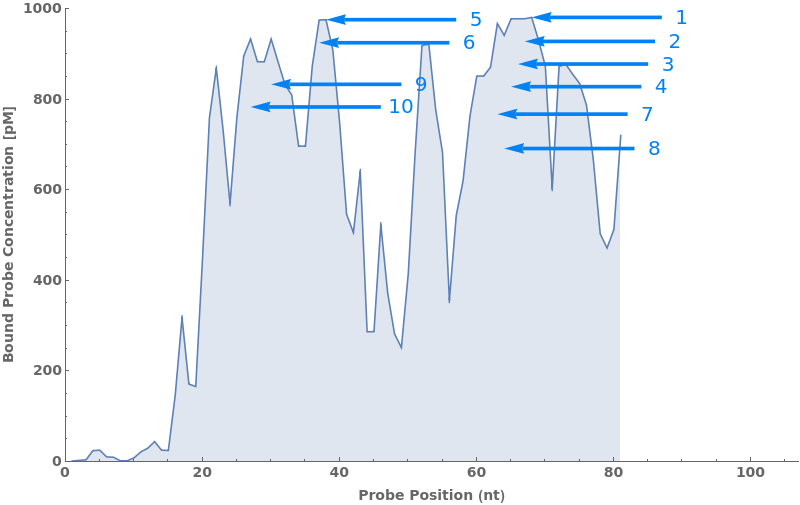
- Once a mechanism is generated at a site a kinetic simulation is performed (via SimulateKinetics) to calculate the concentration of the probe correctly bound to the desired target site.
- The final returned result is a list of probe sequences (one for each site tested) sorted by their correctly bound concentration as determined by the kinetic simulation. Therefore, the first returned probe sequence is the one that is predicted to most faithfully bind to its intended target site.
- Thermodynamic DNA Nearest Neighbor parameters from Object[Report, Literature, "id:kEJ9mqa1Jr7P"]: Allawi, Hatim T., and John SantaLucia. "Thermodynamics and NMR of internal GT mismatches in DNA." Biochemistry 36.34 (1997): 10581-10594.
- Thermodynamic RNA Nearest Neighbor parameters from Object[Report, Literature, "id:M8n3rxYAnNkm"]: Xia, Tianbing, et al. "Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs." Biochemistry 37.42 (1998): 14719-14735.
-
ProbeFoldFalse ProbeFolding is False but Beacon length is set to be nonzero. Please set ProbeFolding to be True or set BeaconStemLength to be zero according to the experimental requirements. ProbeShort ProbeLength (`1`) cannot be smaller than MinPairLevel (`2`). Please increase ProbeLength or decrease MinPairLevel SiteShort At least one of the target sites has a length shorter than the MinPairLevel (`1`). Please increase the probe length.
Input

Output

General Options
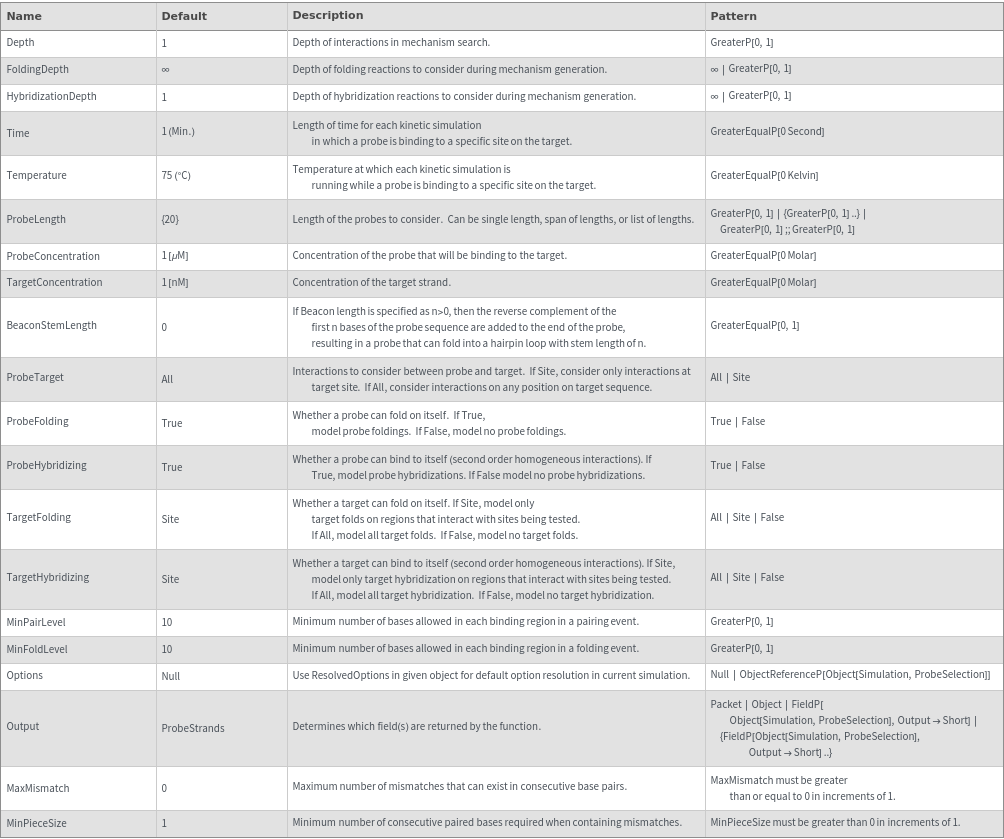
Messages
Examples
Basic Examples (4)
Test every site on the target sequence to find the best probe binding site, probe trands (ProbeStrands field) are returned by defauled sorted by the bounded concentrations in descending order:

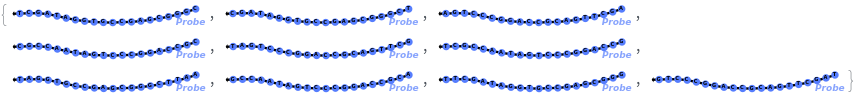
Input can also be a valid nucleotide string:
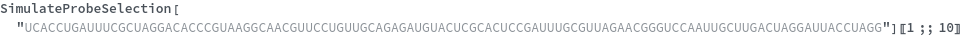

Test only on the explicitly specified sites on the target:

Can be applied directly on a specific ribonuleic acid (RNA) transcript to find the best probe binding site: