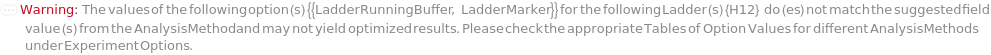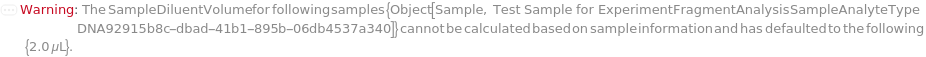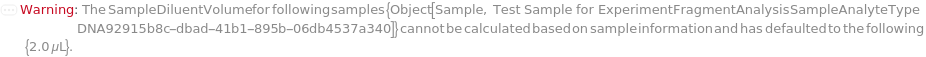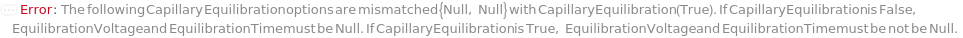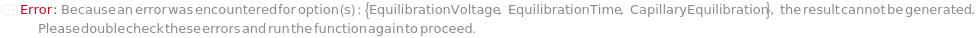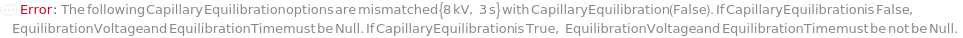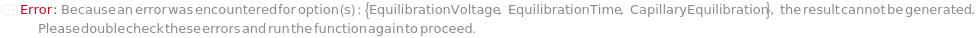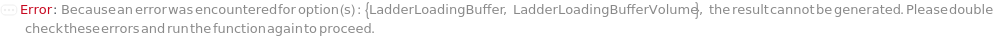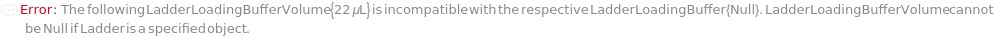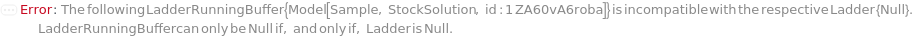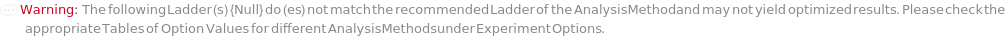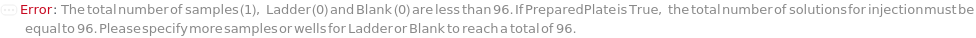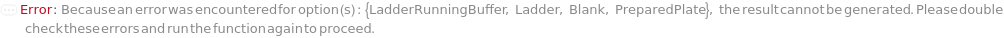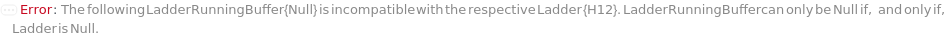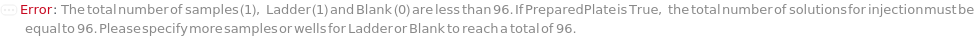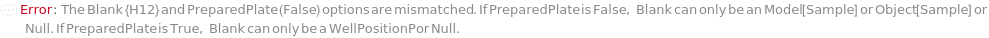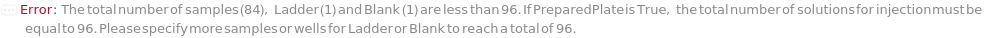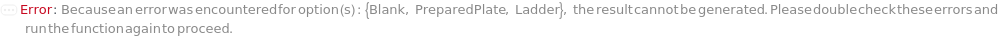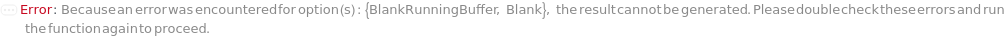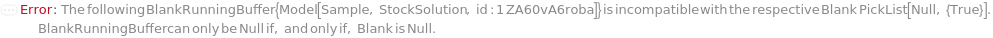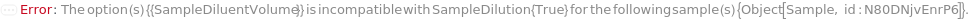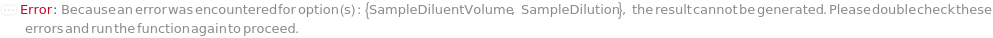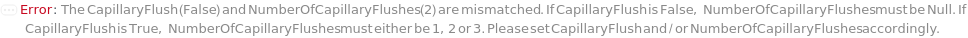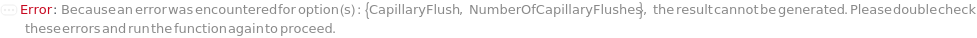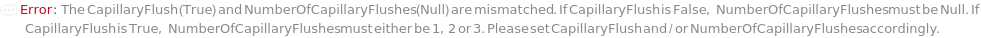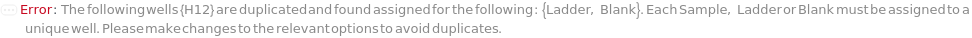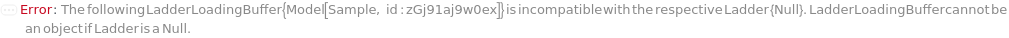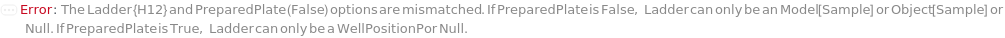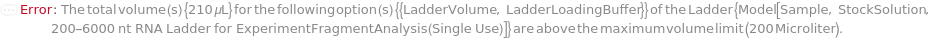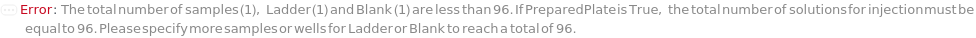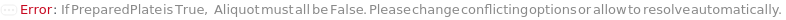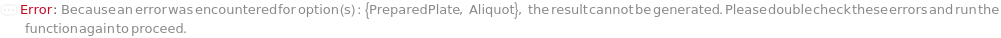General
Instrument
The array-based capillary electrophoresis instrument used for parallel qualitative or quantitative analysis of nucleic acids via separation based on analyte fragment size of up to 84 samples in a single run.
Pattern Description: An object of type or subtype Model[Instrument, FragmentAnalyzer] or Object[Instrument, FragmentAnalyzer]
Programmatic Pattern: ObjectP[{Model[Instrument, FragmentAnalyzer], Object[Instrument, FragmentAnalyzer]}] | Automatic
NumberOfCapillaries
The number of extremely thin, hollow tubes per bundle in the CapillaryArray used in the course of the experiment. Determines the maximum number of samples ran in parallel in a single injection, including a ladder sample which is recommended for every run.
Default Calculation: If the CapillaryArray is specified, automatically set to the NumberOfCapillaries field of the CapillaryArray option. Otherwise, automatically set based on the number of SamplesIn, Ladders (if specified) and Blank (if specified).
Programmatic Pattern: FragmentAnalyzerNumberOfCapillariesP | Automatic
CapillaryArrayLength
The length (Short) of the extremely thin hollow tubes (capillary arrays) that is used by the instrument for analyte separation. Short capillaries have shorter separation run times, as well as improved peak efficiency and sensitivity, but yield lower separation resolution. Long capillaries have longer run times but are ideal if a higher separation resolution is desired. For a Short capillary array, the length from the sample inlet end until the detector window (EffectiveLength) is 33 Centimeter, while the length from the sample inlet until outlet end to the reservoir (TotalLength) is 55 Centimeter. For an illustration, see Figure 2.1.1 under Instrumentation.
Pattern Description: Short.
Programmatic Pattern: FragmentAnalyzerCapillaryArrayLengthP
CapillaryArray
The ordered bundle of extremely thin, hollow tubes (capillary array) that is used by the instrument for analyte separation. Each CapillaryArray has a specific NumberOfCapillaries (96) which indicates the maximum number of samples that are ran in parallel. Each capillary in the array also has a designated CapillaryArrayLength (Short) which affects the separation run times and resolution. Short capillaries have shorter run times, as well as improved peak efficiency and sensitivity, but yield lower separation resolution, and are used by default. Long capillaries have longer run times but are ideal if a higher separation resolution is desired.
Default Calculation: If NumberOfCapillaries is set to 96, set to Model[Part, CapillaryArray, "96-Capillary Array Short"].
Pattern Description: An object of type or subtype Object[Part, CapillaryArray] or Model[Part, CapillaryArray]
Programmatic Pattern: ObjectP[{Object[Part, CapillaryArray], Model[Part, CapillaryArray]}] | Automatic
AnalysisMethod
The method object that contains a set of option values optimized and recommended by the instrument manufacturer that is used as a template for the parameters of the experiment. If not specified by User, an AnalysisMethod is selected from a list of methods for either standard dsDNA or RNA samples according to a selection criteria based on AnalysisStrategy, CapillaryArrayLength and sample information: type of analyte (DNA or RNA), analyte concentrations (High Sensitiviy (Picogram/Microliter) or Standard Sensitivity (Nanogram/Microliter)) and fragment size range (ReadLength, in base pairs or number of nucleotides) of the sample. For more specialized samples (eg DNA smears, plasmid DNA, cfDNA, NGS libraries, samples for CRISPR/Cas9, genomic DNA), it is recommended that AnalysisMethod is selected and specified. For a selection criteria guide on determining the appropriate AnalysisMethod, refer to Figure 3.1 under AnalysisMethod of Experiment Options. Default option values for each AnalysisMethod, including reagents (SeparationGel, LoadingBuffer, Ladder, Blank, Marker, PreMarkerRinseBuffer, PreSampleRinseBuffer), as well as specific parameters of the experiment categories (eg. Sample Preparation, Capillary Conditioning, Capillary Equilibration, Pre-Marker Rinse, Marker Injection, Pre-Sample Rinse, Sample Injection, Separation) are specified in fields of the various Object[Method, FragmentAnalysis] objects available.
Default Calculation: Automatically set to appropriate Object[Method,FragmentAnalysis] based on AnalysisStrategy, CapillaryArrayLength and sample information: type of analyte (equal to TargetAnalyteType field in the AnalysisMethod), analyte concentration (greater than MinTargetConcentration field and less than MaxTargetConcentration field of AnalysisMethod) and fragment size range (MinReadLength greater than or equal to MinTargetReadLength field and MaxReadLength less than or equal to MaxTargetReadLength field of the AnalysisMethod), if available. For a selection criteria guide on determining the appropriate AnalysisMethod, refer to Figure 3.1 under AnalysisMethod of Experiment Options.
Pattern Description: Agilent Method Objects or Custom Method Objects.
Programmatic Pattern: (ObjectP[{Object[Method, FragmentAnalysis]}] | ObjectP[{Object[Method, FragmentAnalysis]}]) | Automatic
PreparedPlate
Indicates if SamplesIn are in an instrument-compatible plate (Model[Container, Plate, "96-well Semi-Skirted PCR Plate for FragmentAnalysis"]) that contain all other necessary components (Ladders, Blanks, LoadingBuffer (if applicable)). If PreparedPlate is True, Ladders and Blanks, if any, are designated by Well Position(s) that contain ladder or blank solutions. All wells contain a solution of least 24 Microliter volume to avoid damage to the capillary array. Prepared plates are directly placed in the sample drawer of the instrument, ready for injection, and does not involve any plate preparation steps. Options including SampleVolume, SampleDiluent, SampleDiluentVolume, SampleLoadingBuffer, SampleLoadingBufferVolume, LadderVolume, LadderLoadingBuffer and LadderLoadingBufferVolume are not applicable and are set to Null.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
MaxNumberOfRetries
The maximum number of separation runs that can be performed for contents of the sample plate when the raw data (electropherogram) for a sample or ladder indicates no peak(s) detected as assessed by the lack of any baseline-corrected signal above 100 RFU. For each separation run retry, resources such as SeparationGel, Dye and ConditioningSolution are re-prepared by preparing new solutions in new containers. Plate resources (SamplePlate,RunningBufferPlate,PreMarkerRinseBufferPlate and PreSampleRinseBufferPlate) are re-prepared by moving the contents to a new plate with a newly re-assigned well position for any sample(s)/ladder(s) that are affected, as well as their index-matched counterparts (RunningBuffer, PreMarkerRinseBuffer, PreSampleRinseBuffer). The well re-assignment is to avoid the underperforming capillary array assigned in the previous run. Note that the MarkerPlate is not re-prepared and is used as-is for every retry, if applicable, due to the oil-layer that prevents effective reusability of the transferred components.
Pattern Description: Greater than or equal to 1 and less than or equal to 3 or Null.
Programmatic Pattern: RangeP[1, 3] | Null
NumberOfReplicates
The number of wells each input sample is loaded into. For example {input 1, input 2} with NumberOfReplicates->2 will act like {input 1, input 1, input 2, input 2}.
Pattern Description: Greater than or equal to 2 in increments of 1 or Null.
Programmatic Pattern: GreaterEqualP[2, 1] | Null
SampleAnalyteType
The nucleic acid type (DNA or RNA) of the analytes in the sample that are separated based on fragment size.
Default Calculation: Automatically set to the most PolymerType that is either DNA or RNA that can be found under Analytes. If Analytes is not populated, automatically set to the most PolymerType that is either DNA or RNA that can be found under Composition. Otherwise, set to DNA.
Pattern Description: DNA or RNA.
Programmatic Pattern: FragmentAnalysisAnalyteTypeP | Automatic
Index Matches to: experiment samples
MinReadLength
The number (or estimated number) of base pairs or nucleotides of the shortest fragment analyte in the sample.
Default Calculation: Automatically set to the lowest number of base pairs or nucleotides of identity models of PolymerType DNA or RNA under the Composition field of the sample, as determined by SequenceLength. Otherwise, automatically set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 60000 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 60000, 1] | Automatic) | Null
Index Matches to: experiment samples
MaxReadLength
The number (or estimated number) of base pairs or nucleotides of the longest fragment analyte in the sample.
Default Calculation: Automatically set to the highest number of base pairs or nucleotides of identity models of PolymerType DNA or RNA under the Composition field of the sample, as determined by SequenceLength. Otherwise, automatically set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 60000 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 60000, 1] | Automatic) | Null
Index Matches to: experiment samples
AnalysisStrategy
The objective of the analysis of the samples. Qualitative performs quality assessment of samples and outputs include gel image, electropherogram, total concentration and sample quality number (DNA Quality Number (DQN) or RNA Quality Number (RQN)). Quantitative performs everything a Qualitative analysis does, with the exception of the default loading buffer added to the sample(s) or ladder(s) containing quantified markers that serve as internal standard and allows measurement analyte concentration with better accuracy than Qualitative methods.
Default Value: Qualitative
Pattern Description: Qualitative or Quantitative.
Programmatic Pattern: FragmentAnalysisStrategyP
Method Saving
AnalysisMethodName
The name that is given to the Object[Method,FragmentAnalysis] that is generated from options specified in the experiment and is uploaded after the experiment is done. Note that if no AnalysisMethodName is provided, no Object[Method,FragmentAnalysis] is saved.
Pattern Description: A string or Null.
Programmatic Pattern: _String | Null
Sample Preparation
SampleVolume
The volume of sample (after aliquoting, if applicable) that is transferred into the plate, prior to addition of SampleDiluent (if applicable) and SampleLoadingBuffer (if applicable), and before loading into the instrument. Each sample is dispensed from Position A1 onwards (from A1, A2, A3...). If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: If SampleDilution is False, automatically set to the TargetSampleVolume field of the AnalysisMethod. If SampleDilution is True, automatically determined based on the concentration of the first identity model of PolymerType DNA or RNA under Composition, and the MaxTargetMassConcentration field of the AnalysisMethod. Otherwise, automatically set to 2 Microliter. If PreparedPlate is True, automatically set to Null.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SampleDilution
Indicates if SampleDiluent is added to the sample (after aliquoting, if applicable) to reduce the sample concentration prior to addition of the SampleLoadingBuffer and loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
Index Matches to: experiment samples
SampleDiluent
The buffer solution added to the sample to reduce the sample concentration prior to addition of the SampleLoadingBuffer and loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: If SampleDilution is set to True, automatically set to Model[Sample, "1x Tris-EDTA (TE) Buffer for ExperimentFragmentAnalysis"]. If PreparedPlate is True OR SampleDilution is set to False, automatically set to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
SampleDiluentVolume
The volume of buffer solution added to the sample to reduce the sample concentration prior to addition of the SampleLoadingBuffer and loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: If SampleDilution is set to True, automatically determined based on the concentration of the identity models of PolymerType matching SampleAnalyteType under Composition, and the MaxTargetMassConcentration field of the AnalysisMethod. Otherwise, automatically set to the difference between SampleVolume and TargetSampleVolume. If PreparedPlate is True, automatically set to Null.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SampleLoadingBuffer
The solution added to the sample (after aliquoting, if applicable), to either add markers (Quantitative) or further dilute (Qualitative) the sample prior to loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: Automatically set to the SampleLoadingBuffer field of the AnalysisMethod. If PreparedPlate is True, automatically set to Null. For a list of the default SampleLoadingBuffer for each AnalysisMethod, see Figure 3.2 under SampleLoadingBuffer of Experiment Options.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
SampleLoadingBufferVolume
The volume of SampleLoadingBuffer added to the sample (after aliquoting, if applicable), to either add markers to (Quantitative) or further dilute (Qualitative) the sample prior to loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: Automatically set to the SampleLoadingBufferVolume field of the AnalysisMethod. If PreparedPlate is True, automatically set to Null. For a list of the default SampleLoadingBufferVolume for each AnalysisMethod, see Figure 3.2 under SampleLoadingBuffer of Experiment Options.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
Ladder
The solution(s) that contain nucleic acids of known lengths used for qualitative or quantitative data analysis. One ladder solution, which is dispensed to Position H12 (for a 96-capillary array), is recommended for each run of the capillary array. If multiple ladders are specified, each ladder is dispensed on the sample plate to occupy successive wells up until Position H12 (for a 96-capillary array). For example, with a 96-capillary array with 3 specified ladders, the first ladder is dispensed to Position H10, the second ladder to Position H11 and the third ladder to position H12. If PreparedPlate is set to True, Ladder is specified as the Well Position(s) the ladder solution(s) has been dispensed to, and are consecutive Well Positions to end in H12 (for example, if there are three ladder solutions in the PreparedPlate, Ladder is set to H10, H11 and H12).
Default Calculation: If PreparedPlate is False, automatically set to the Ladder field of the AnalysisMethod and is dispensed to occupy successive wells until Position H12 for a 96-capillary array. If PreparedPlate is True, automatically set to {"H12"} unless Well Position(s) are specified. For a list of the default Ladders for each AnalysisMethod, see Figure 3.3 under Ladder of Experiment Options. At ladder is always required at Position H12.
Pattern Description: Ladder Solution or Prepared Plate Well or Null.
Programmatic Pattern: (((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Alternatives @@ Flatten[AllWells[NumberOfWells -> 96]]) | Automatic) | Null
LadderVolume
The volume of ladder that is transferred into the plate, prior to addition of LadderLoadingBuffer, and before loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: Automatically set to the LadderVolume field of the AnalysisMethod. If PreparedPlate is True, automatically set to Null. For a list of the default LadderVolume for each AnalysisMethod, see Figure 3.3 under Ladder of Experiment Options.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
LadderLoadingBuffer
The solution added to the Ladder, to either add markers to or further dilute the Ladder prior to loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: Automatically set to the LadderLoadingBuffer field of the AnalysisMethod. If PreparedPlate is True, automatically set to Null. For a list of the default LadderLoadingBuffer for each AnalysisMethod, see Figure 3.4 under LadderLoadingBuffer of Experiment Options.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
LadderLoadingBufferVolume
The volume of LadderLoadingBuffer added to the Ladder (after aliquoting, if applicable), to either add markers to or further dilute the Ladder prior to loading into the instrument. If PreparedPlate is True, this option is not applicable and is set to Null.
Default Calculation: Automatically set to the LadderLoadingBufferVolume field of the AnalysisMethod. If PreparedPlate is True, automatically set to Null. For a list of the default LoadingBufferVolume for each AnalysisMethod, see Figure 3.4 under LadderLoadingBuffer of Experiment Options.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[1*Microliter, 200*Microliter] | Automatic) | Null
Blank
The solution that is dispensed in the well(s) of the sample plate required to be filled that are not filled by a sample or a ladder. For example, for a run using a 96-capillary array, if there are only 79 samples and 1 ladder, 16 wells are filled with Blank.). Wells filled with Blank each contain a volume equal to the total volume of TargetSampleVolume and LoadingBufferVolume of the AnalysisMethod. If PreparedPlate is set to True, Blank is specified as the Well Position(s) the blank solution(s) have been dispensed to.
Default Calculation: Automatically set to the Blank field of the AnalysisMethod if there are wells unoccupied by the samples or ladders. If PreparedPlate is True and Blank is not specified, automatically set to Null. For a list of the default Blank for each AnalysisMethod, see Figure 3.5 under Blank of Experiment Options.
Pattern Description: Blank Solution or Prepared Plate Well or Null.
Programmatic Pattern: (((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Alternatives @@ Flatten[AllWells[NumberOfWells -> 96]]) | Automatic) | Null
Capillary Conditioning
SeparationGel
The gel reagent that serves as sieving matrix to facilitate the optimal separation of the analyte fragments in each sample by size. Each capillary in the array is flushed with the ConditioningSolution followed by filling with a mixture of the SeparationGel and the Dye prior to sample runs.
Default Calculation: Automatically set to the SeparationGel field of the AnalysisMethod. For a list of the default SeparationGel for each AnalysisMethod, see Figure 3.6 under SeparationGel of Experiment Options.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic
Dye
The solution of a dye molecule that fluoresces when bound to DNA or RNA fragments in the sample and is used in the detection of the analyte fragments as it passes through the detection window of the capillary. Each capillary in the array is flushed with the ConditioningSolution followed by filling with a mixture of the SeparationGel and the Dye prior to sample runs.
Default Calculation: Automatically set to the Dye field of the AnalysisMethod. The default Dye for most AnalysisMethod is Model[Sample, "Intercalating Dye for ExperimentFragmentAnalysis"]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic
ConditioningSolution
The solution used in the priming of the capillaries before filling with the SeparationGel and Dye prior to a sample run. The flushing of the capillary array with the conditioning solution restores capillary performance and helps maintain the low and reproducible electroosmotic flow for more precise analyte mobilities and migration times by stabilizing buffer pH and background electrolyte levels. This step also acts to flush out unwanted molecules or reagents from previous runs to minimize contamination of samples.
Default Calculation: Automatically set to the ConditioningSolution field of the AnalysisMethod. The default conditioning solution for most AnalysisMethod is Model[Sample, StockSolution, "1X Conditioning Solution for ExperimentFragmentAnalysis"]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic
Capillary Flush
CapillaryFlush
Indicates if an optional CapillaryFlush procedure is performed prior to a sample run. A CapillaryFlush step involves running specified CapillaryFlushSolution(s) through the capillaries and into a Waste Plate. CapillaryFlush is False by default since a capillary conditioning step (which also has the effect of washing the capillaries) is always performed prior to every sample run.
Default Calculation: If NumberOfCapillaries is set to a number between 1 to 3, automatically set to True. Otherwise, set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
NumberOfCapillaryFlushes
The number of CapillaryFlush steps that are performed prior to the sample run, where the specified CapillaryFlushSolution for each step runs through the capillaries and into the Waste Plate.
Default Calculation: If CapillaryFlush is set to True, automatically set to 1.
Pattern Description: Greater than or equal to 1 and less than or equal to 3 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 3, 1] | Automatic) | Null
PrimaryCapillaryFlushSolution
The solution that runs through the capillaries and into the Waste Plate during the first CapillaryFlush step.
Default Calculation: If CapillaryFlush is set to True, automatically set to Model[Sample, StockSolution, "1X Conditioning Solution for ExperimentFragmentAnalysis"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
PrimaryCapillaryFlushPressure
The positive pressure applied at the capillaries' destination by a high pressure syringe pump that drives the PrimaryCapillaryFlushSolution through the capillaries backwards from the reservoir and into the Waste Plate during the first CapillaryFlush step.
Default Calculation: If CapillaryFlush is set to True, automatically set to 280 PSI.
Pattern Description: Greater than or equal to 0.1 pounds
‐force per inch squared and less than or equal to 300 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[0.1*PSI, 300*PSI] | Automatic) | Null
PrimaryCapillaryFlushFlowRate
The flow rate of the PrimaryCapillaryFlushSolution as it runs through the capillaries and into the Waste Plate during the first CapillaryFlush step.
Default Calculation: If CapillaryFlush is set to True, automatically set to 200 Microliter/Second.
Pattern Description: Greater than or equal to 1 microliter per second and less than or equal to 1000 microliters per second or Null.
Programmatic Pattern: (RangeP[1*(Microliter/Second), 1000*(Microliter/Second)] | Automatic) | Null
PrimaryCapillaryFlushTime
The duration for which the PrimaryCapillaryFlushSolution runs through the capillaries and into the Waste Plate during the first CapillaryFlush step.
Default Calculation: If CapillaryFlush is set to True, automatically set to 180 Second.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 240 minutes or Null.
Programmatic Pattern: (RangeP[1*Minute, 240*Minute] | Automatic) | Null
SecondaryCapillaryFlushSolution
The solution that runs through the capillaries and into the Waste Plate during the second CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is greater than 1, automatically set to Model[Sample,"Agilent Conditioning Solution for Fragment Analysis"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
SecondaryCapillaryFlushPressure
The positive pressure applied at the capillaries' destination by a high pressure syringe pump that drives the SecondaryCapillaryFlushSolution through the capillaries backwards from the reservoir and into the Waste Plate during the second CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is greater than 1, automatically set to 280 PSI.
Pattern Description: Greater than or equal to 1 pound
‐force per inch squared and less than or equal to 300 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[1*PSI, 300*PSI] | Automatic) | Null
SecondaryCapillaryFlushFlowRate
The flow rate of the SecondaryCapillaryFlushSolution as it runs through the capillaries and into the Waste Plate during the second CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is greater than 1, automatically set to 200 Microliter/Second.
Pattern Description: Greater than or equal to 1 microliter per second and less than or equal to 1000 microliters per second or Null.
Programmatic Pattern: (RangeP[1*(Microliter/Second), 1000*(Microliter/Second)] | Automatic) | Null
SecondaryCapillaryFlushTime
The duration for which the SecondaryCapillaryFlushSolution runs through the capillaries and into the Waste Plate during the second CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is greater than 1, automatically set to 180 Second.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 240 minutes or Null.
Programmatic Pattern: (RangeP[1*Minute, 240*Minute] | Automatic) | Null
TertiaryCapillaryFlushSolution
The solution that runs through the capillaries and into the Waste Plate during the third CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is 3, automatically set to Model[Sample,"Agilent Conditioning Solution for Fragment Analysis"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
TertiaryCapillaryFlushPressure
The positive pressure applied at the capillaries' destination by a high pressure syringe pump that drives the TertiaryCapillaryFlushSolution through the capillaries backwards from the reservoir and into the Waste Plate during the third CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is 3, automatically set to 280 PSI.
Pattern Description: Greater than or equal to 1 pound
‐force per inch squared and less than or equal to 300 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[1*PSI, 300*PSI] | Automatic) | Null
TertiaryCapillaryFlushFlowRate
The flow rate of the TertiaryCapillaryFlushSolution as it runs through the capillaries and into the Waste Plate during the third CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is 3, automatically set to 200 Microliter/Second.
Pattern Description: Greater than or equal to 1 microliter per second and less than or equal to 1000 microliters per second or Null.
Programmatic Pattern: (RangeP[1*(Microliter/Second), 1000*(Microliter/Second)] | Automatic) | Null
TertiaryCapillaryFlushTime
The duration for which the TertiaryCapillaryFlushSolution runs through the capillaries and into the Waste Plate during the third CapillaryFlush step.
Default Calculation: If NumberOfCapillaryFlushes is 3, automatically set to 180 Second.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 240 minutes or Null.
Programmatic Pattern: (RangeP[1*Minute, 240*Minute] | Automatic) | Null
Capillary Equilibration
CapillaryEquilibration
Indicates if a voltage is run through the capillaries as the source tips of the capillaries are immersed in the wells of the RunningBufferPlate in order to prepare the gel inside the capillaries and normalize the electroosmotic flow for more precise analyte mobilities and migration times during separation. If applicable, this is done after the introduction of the SeparationGel and Dye into the capillaries.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
PreparedRunningBufferPlate
The pre-prepared 96-well plate which contains the buffer solutions that help conduct current through the gel in the capillaries and facilitates the separation of analyte fragments. The tips of the capillaries are immersed in the wells of the RunningBufferPlate during the CapillaryEquilibration and Separation steps. The contents of the wells matching the well position of the Samples, Ladder(s) and Blank(s) serve as the SampleRunningBuffer, LadderRunningBuffer (if applicable) and BlankRunningBuffer (if applicable). In order for the capillary tips to reach the solution, a minimum volume of 1 Milliliter of buffer solution is required in the each well of the plate and must be in plate model Model[Container, Plate, "96-well 1mL Deep Well Plate (Short) for FragmentAnalysis"].
Pattern Description: An object of type or subtype Object[Container, Plate] or a prepared sample or Null.
Programmatic Pattern: (ObjectP[Object[Container, Plate]] | _String) | Null
SampleRunningBuffer
The buffer solution that contains ions that help conduct current through the gel in the capillaries and facilitates the separation of analyte fragments in the sample. The tips of the capillaries are immersed in the wells of a RunningBufferPlate during the CapillaryEquilibration and Separation steps. If PreparedRunningBufferPlate is not specified, a new 96-well plate that contains the SampleRunningBuffer in the matching WellPosition(s) of the SamplesIn is prepared.
Default Calculation: If PreparedRunningBufferPlate is specified, set to the content(s) of the well matching the well position of the SamplesIn. If PreparedRunningBufferPlate is Null, automatically set to the SampleRunningBuffer field of the AnalysisMethod. Otherwise, automatically set to Model[Sample, StockSolution, "1x Running Buffer for ExperimentFragmentAnalysis"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic
Index Matches to: experiment samples
LadderRunningBuffer
The buffer solution that contains ions that help conduct current through the gel in the capillaries and facilitates the separation of fragments in the ladder. The tips of the capillaries are immersed in the wells of a RunningBufferPlate during the CapillaryEquilibration and Separation steps. If PreparedRunningBufferPlate is not specified, a new 96-well plate that contains the LadderRunningBuffer in the matching WellPosition(s) of the Ladder is prepared.
Default Calculation: If PreparedRunningBufferPlate is specified and Ladder is not Null, set to the content(s) of the well matching the well position of the Ladder(s). If PreparedRunningBufferPlate is Null and Ladder is not Null, automatically set to the LadderRunningBuffer field of the AnalysisMethod.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankRunningBuffer
The buffer solution that contains ions that help conduct current through the gel in the capillaries that contain the blank. The tips of the capillaries are immersed in the wells of a RunningBufferPlate during the CapillaryEquilibration and Separation steps. If PreparedRunningBufferPlate is not specified, a new 96-well plate that contains the BlankRunningBuffer in the matching WellPosition(s) of the Blank is prepared.
Default Calculation: If PreparedRunningBufferPlate is specified and Blank is not Null, set to the content(s) of the well matching the well position of the Blank(s). If PreparedRunningBufferPlate is Null and Blank is not Null, automatically set to the BlankRunningBuffer field of the AnalysisMethod.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
RunningBufferPlateStorageCondition
The non-default condition under which the RunningBufferPlate is stored after the protocol is completed.
Default Value: Refrigerator
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal.
Programmatic Pattern: SampleStorageTypeP | Disposal
EquilibrationVoltage
The electric potential applied across the capillaries as the capillaries are immersed in the RunningBuffer to condition the gel inside the capillaries and normalize the electroosmotic flow for more precise analyte mobilities and migration times prior to a Pre-Marker Rinse or a Pre-Sample Rinse step.
Default Calculation: If CapillaryEquilibration is set to False, automatically set to Null. If CapillaryEquilibration is set to True and EquilibrationVoltage field of the AnalysisMethod is not Null, set to the EquilibrationVoltage field of the AnalysisMethod. If CapillaryEquilibration is set to True and EquilibrationVoltage field of the AnalysisMethod is Null, set to the 8.0 Kilovolt.
Pattern Description: Greater than or equal to 0 kilovolts and less than or equal to 15 kilovolts or Null.
Programmatic Pattern: (RangeP[0*Kilovolt, 15*Kilovolt] | Automatic) | Null
EquilibrationTime
The duration for which electric potential (EquilibrationVoltage) is applied across the capillaries as the capillaries are immersed in the RunningBuffer to condition the gel inside the capillaries and normalize the electroosmotic flow for more precise analyte mobilities and migration times.
Default Calculation: If CapillaryEquilibration is True and AnalysisMethod is specified, automatically set to the EquilibrationTime field of the AnalysisMethod.
Pattern Description: Greater than or equal to 1 second and less than or equal to 20 minutes or Null.
Programmatic Pattern: (RangeP[1*Second, 20*Minute] | Automatic) | Null
Pre-Marker Rinse
PreMarkerRinse
Indicates if the tips of the capillaries are rinsed by dipping them in and out of the PreMarkerRinseBuffer contained in the wells of a compatible 96-well plate in order to wash off any previous reagents the capillaries may have come in contact with. This step precedes the MarkerInjection step.
Default Calculation: If the AnalysisMethod is specified, automatically set to the PreMarkerRinse field of the AnalysisMethod. Otherwise, automatically set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
NumberOfPreMarkerRinses
The number of dips of the capillary tips in and out of the PreMarkerRinseBuffer contained in the wells of a compatible 96-well plate prior to MarkerInjection in order to wash off any previous reagents the capillaries may have come in contact with. This step precedes the MarkerInjection step.
Default Calculation: If PreMarkerRinse is True, automatically set to the NumberOfPreMarkerRinses field of the AnalysisMethod. If PreMarkerRinse is True and the NumberOfPreMarkerRinses field of the AnalysisMethod is Null, automatically set to 1. If PreMarkerRinse is False, automatically set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 20 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 20, 1] | Automatic) | Null
PreMarkerRinseBuffer
The buffer solution that is used to rinse the capillary tips by dipping them in and out of the PreMarkerRinseBuffer contained in the wells of a compatible 96-well plate in order to wash off any previous reagents the sample capillaries may have come in contact with. This step precedes the MarkerInjection step.
Default Calculation: If PreMarkerRinse is True, automatically set to the PreMarkerRinseBuffer field of the AnalysisMethod. For a list of the default PreMarkerRinseBuffer for each AnalysisMethod, see Figure 3.8 under PreMarkerRinse of Experiment Options.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
PreMarkerRinseBufferPlateStorageCondition
The non-default condition under which the PreMarkerRinseBuffer (contained in the PreMarkerRinseBufferPlate) is stored after the protocol is completed.
Default Calculation: If PreMarkerRinseBuffer is not Null, automatically set to Disposal. Otherwise, set to Null.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Marker Injection
MarkerInjection
Indicates if injection of SampleMarker, LadderMarker (if applicable) and BlankMarker (if applicable) into the capillaries is performed prior to a sample run. SampleMarker, LadderMarker and BlankMarker are solutions contained in a MarkerPlate that contains upper and/or lower marker that elutes at a time corresponding to a known nucleotide size.
Default Calculation: If the AnalysisMethod is specified, automatically set to the MarkerInjection field of the AnalysisMethod. Otherwise, automatically set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
PreparedMarkerPlate
The pre-prepared 96-well plate which contains the marker solutions that contain upper and/or lower marker that elutes at a time corresponding to a known nucleotide size. The contents of the wells matching the well position of the Samples, Ladder(s) and Blank(s) serve as the SampleMarker, LadderMarker (if applicable) and BlankMarker (if applicable). In order for the capillary tips to reach the solution, a minimum volume of 50 Microliter of solution is required in the each well of the plate.
Pattern Description: An object of type or subtype Object[Container, Plate] or a prepared sample or Null.
Programmatic Pattern: (ObjectP[Object[Container, Plate]] | _String) | Null
SampleMarker
The solution that contains upper and/or lower marker that elutes at a time corresponding to a known nucleotide size. Tha SampleMarker is injected into the capillaries during MarkerInjection and runs with the samples during Separation. If PreparedMarkerPlate is not specified, a new 96-well plate that contains the SampleMarker in the matching WellPosition(s) of the SamplesIn is prepared.
Default Calculation: If PreparedMarkerPlate is specified, set to the content(s) of the well matching the WellPosition of the SamplesIn. If PreparedMarkerPlate is Null and MarkerInjection is True, automatically set to the SampleMarker field of the AnalysisMethod, if not Null. If PreparedMarkerPlate is Null and MarkerInjection is True and SampleMarker field of AnalysisMethod is Null, automatically set to Model[Sample, "35 bp and 5000 bp Markers for ExperimentFragmentAnalysis"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
LadderMarker
The solution that contains upper and/or lower marker that elutes at a time corresponding to a known nucleotide size. Tha LadderMarker is injected into the capillaries during MarkerInjection and runs with the Ladder(s) during Separation. If PreparedMarkerPlate is not specified, a new 96-well plate that contains the LadderMarker in the matching WellPosition(s) of the Ladder(s) is prepared.
Default Calculation: If PreparedMarkerPlate is specified, set to the content(s) of the well matching the WellPosition of the Ladder. If PreparedMarkerPlate is Null, MarkerInjection is True and Ladder is not Null, automatically set to the LadderMarker field of the AnalysisMethod, if not Null. If PreparedMarkerPlate is Null, MarkerInjection is True, Ladder is not Null and LadderMarker field of AnalysisMethod is Null, automatically set to Model[Sample, "100-3000 bp DNA Ladder for ExperimentFragmentAnalysis"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankMarker
The solution that contains upper and/or lower marker that elutes at a time corresponding to a known nucleotide size. Tha BlankMarker is injected into the capillaries during MarkerInjection and runs with the Blank(s) during Separation. If PreparedMarkerPlate is not specified, a new 96-well plate that contains the BlankMarker in the matching WellPosition(s) of the Blank(s) is prepared.
Default Calculation: If PreparedMarkerPlate is specified, set to the content(s) of the well matching the WellPosition of the Blank. If PreparedMarkerPlate is Null, MarkerInjection is True and Blank is not Null, automatically set to the BlankMarker field of the AnalysisMethod, if not Null. If PreparedMarkerPlate is Null, MarkerInjection is True, Blank is not Null and BlankMarker field of AnalysisMethod is Null, automatically set to Model[Sample, "100-3000 bp DNA Ladder for ExperimentFragmentAnalysis"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
MarkerInjectionTime
The duration an electric potential (VoltageInjection) is applied across the capillaries to drive the Marker into the capillaries. In VoltageInjection, the injected volume is proportional to the MarkerInjectionTime. In addition, a short InjectionTime reduces band broadening while a longer InjectionTime can serve to minimize voltage or pressure variability during injection.
Default Calculation: If the AnalysisMethod is specified, automatically set to the MarkerInjectionTime field of the AnalysisMethod. If MarkerInjection is True and AnalysisMethod is not specified, automatically set to 5 Second. For a list of the default MarkerInjectionTime for each AnalysisMethod, see Figure 3.9 under MarkerInjection of Experiment Options.
Pattern Description: Greater than or equal to 1 second and less than or equal to 20 minutes or Null.
Programmatic Pattern: (RangeP[1*Second, 20*Minute] | Automatic) | Null
MarkerInjectionVoltage
The electric potential applied across the capillaries to drive the Marker forward into the capillaries, from the Marker Plate to the capillary inlet, during the MarkerInjection step. The injected volume is proportional to the MarkerInjectionVoltage, where the higher the MarkerInjectionVoltage, the higher the injected volume. For a list of the default MarkerInjectionVoltage for each AnalysisMethod, see Figure 3.9 under MarkerInjection of Experiment Options.
Default Calculation: If the AnalysisMethod is specified, automatically set to the MarkerInjectionVoltage field of the AnalysisMethod. If MarkerInjection is True, OR AnalysisMethod is not specified, set to 3 Kilovolt.
Pattern Description: Greater than or equal to 0 kilovolts and less than or equal to 149.7 kilovolts or Null.
Programmatic Pattern: (RangeP[0*Kilovolt, 149.7*Kilovolt] | Automatic) | Null
MarkerPlateStorageCondition
The non-default condition under which the MarkerPlate is stored after the protocol is completed.
Default Calculation: If SampleMarker, LadderMarker OR BlankMarker is Not Null, automatically set to Refrigerator. Otherwise, set to Null.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Pre-Sample Rinse
PreSampleRinse
Indicates if the tips of the capillaries are rinsed by dipping them in and out of the PreSampleRinseBuffer contained in the wells of a compatible 96-well plate in order to wash off any previous reagents the capillaries may have come in contact with. This step precedes the sampleInjection step.
Default Calculation: If the AnalysisMethod is specified, automatically set to the PreSampleRinse field of the AnalysisMethod. Otherwise, automatically set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
NumberOfPreSampleRinses
The number of dips of the capillary tips in and out of the PreSampleRinseBuffer contained in the wells of a compatible 96-well plate prior to MarkerInjection in order to wash off any previous reagents the capillaries may have come in contact with. This step precedes the SampleInjection step.
Default Calculation: If PreSampleRinse is True, automatically set to the NumberOfPreSampleRinses field of the AnalysisMethod. If PreSampleRinse is True and the NumberOfPreSampleRinses field of the AnalysisMethod is Null, automatically set to 1. If PreSampleRinse is False, automatically set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 20 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 20, 1] | Automatic) | Null
PreSampleRinseBuffer
The buffer solution that is used to rinse the capillary tips by dipping them in and out of the PreSampleRinseBuffer contained in the wells of a compatible 96-well plate in order to wash off any previous reagents the sample capillaries may have come in contact with. This step precedes the SampleInjection step. The recommended buffer used for rinsing (Tris-EDTA buffer) maintains the ideal pH for nucleic acid solubilization (Tris buffer), as well as protects the analyte from degradation by DNAses/RNAses (EDTA).
Default Calculation: If PreSampleRinse is True, automatically set to the PreSampleRinseBuffer field of the AnalysisMethod. For a list of the default PreSampleRinseBuffer for each AnalysisMethod, see Figure 3.10 under PreSampleRinse of Experiment Options.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
PreSampleRinseBufferPlateStorageCondition
The non-default condition under which the PreSampleRinseBuffer (contained in the Pre-Sample Rinse Plate) is stored after the protocol is completed.
Default Calculation: If PreSampleRinseBuffer is not Null, automatically set to Disposal. Otherwise, set to Null.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Sample Injection
SampleInjectionTime
The duration a electric potential (VoltageInjection) is applied across the capillaries to drive the samples into the capillaries. The injected volume is proportional to the SampleInjectionTime. In addition, a short InjectionTime reduces band broadening while a longer InjectionTime can serve to minimize voltage or pressure variability during injection.
Default Calculation: Automatically set to the SampleInjectionTime field of the AnalysisMethod. For a list of the default SampleInjectionTime for each AnalysisMethod, see Figure 3.11 under SampleInjectionTime of Experiment Options.
Pattern Description: Greater than or equal to 1 second and less than or equal to 20 minutes.
Programmatic Pattern: RangeP[1*Second, 20*Minute] | Automatic
SampleInjectionVoltage
The electric potential applied across the capillaries to drive the samples or ladders forward into the capillaries, from the Sample Plate to the capillary inlet, during the SampleInjection step. The injected volume is proportional to the SampleInjectionVoltage, where the higher the SampleInjectionVoltage, the higher the injected volume.
Default Calculation: Automatically set to the SampleInjectionVoltage field of the AnalysisMethod. For a list of the default SampleInjectionVoltage for each AnalysisMethod, see Figure 3.12 under SampleInjectionVoltage of Experiment Options.
Pattern Description: Greater than or equal to 0 kilovolts and less than or equal to 149.7 kilovolts.
Programmatic Pattern: RangeP[0*Kilovolt, 149.7*Kilovolt] | Automatic
Separation
SeparationTime
The duration for which the SeparationVoltage is applied across the capillaries in order for migration of analytes to occur. There should be sufficient SeparationTime for the analytes to migrate from the capillary inlet end to the capillary outlet end for a complete analysis. The higher the SeparationVoltage, the shorter the SeparationTime necessary for complete migration of analytes through the capillaries.
Default Calculation: Automatically set to the SeparationTime field of the AnalysisMethod. For a list of the default SeparationTime for each AnalysisMethod, see Figure 3.13 under SeparationTime of Experiment Options.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 240 minutes.
Programmatic Pattern: RangeP[1*Minute, 240*Minute] | Automatic
SeparationVoltage
The electric potential applied across the capillaries as the sample analytes migrate through the capillaries during the sample run. Higher SeparationVoltage results in faster separations and higher separation efficiencies but can also cause overheating of sample solutions if set too high.
Default Calculation: Automatically set to the SeparationVoltage field of the AnalysisMethod. For a list of the default SeparationVoltage for each AnalysisMethod, see Figure 3.14 under SeparationVoltage of Experiment Options.
Pattern Description: Greater than or equal to 0 kilovolts and less than or equal to 15 kilovolts.
Programmatic Pattern: RangeP[0*Kilovolt, 15*Kilovolt] | Automatic
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Value: Model[Container, Vessel, 2mL Tube]
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: ObjectP[Model[Container]] | Null
Index Matches to: experiment samples
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer.
Default Value: 1 milliliter
Pattern Description: Count or Count or Mass or Volume or Null.
Programmatic Pattern: (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.]) | Null
Index Matches to: experiment samples