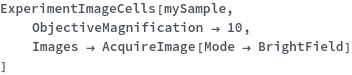
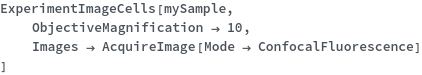
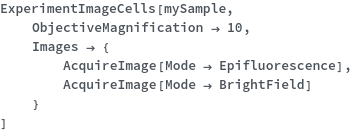
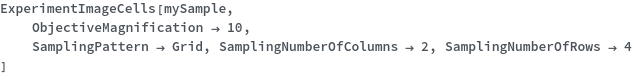ExperimentImageCells
ExperimentImageCells[Samples]⟹Protocol
generates a Protocol for imaging the provided Samples using a widefield microscope or a high content imager. Samples may be imaged in bright-field, phase contrast, epifluorescence, or confocal fluorescence mode.
Conventional optical microscopes use specialized optics to focus light and illuminate samples in order to view structures as small as 200 nm. Several types of microscopes are used to image cells and tissues: bright-field, epifluorescence (widefield), and confocal, with each differing in light sources, optical lenses, and other hardware. Bright-field microscopes are the simplest type of microscope and use white light to illuminate the sample, with denser areas transmitting less light. This microscope is typically used for quick analysis, such as during routine cell maintenance or cell counting. Many bright-field microscopes intended for imaging biological samples also contain phase filters to increase the contrast between the sample and background. Epifluorescence or widefield microscopes are designed to image samples that are labeled with fluorescent compounds and enable more nuanced imaging for assays including colocalization studies and dynamic, live-cell studies. Specialized filters on the microscope selectively pass illumination light at particular wavelength ranges to excite the fluorophores of interest. The emitted fluorescence from the samples is then captured by a digital camera. Common fluorescence filters include those intended to image fluorophores in the blue, green, red, and far-red range. Confocal microscopes contain one or more pinholes in the light path to block out-of-focus light in order to increase optical resolution. Confocal microscopy is often used to image thicker samples to clearly image structures that vary in height along the z-axis. Confocal microscopes are typically equipped with fluorescent filters to image a wide range of fluorophores. Additionally, confocal microscope has been developed into a high-content imager, which enables automated high-throughput imaging of a large number of samples.
Experimental Principles
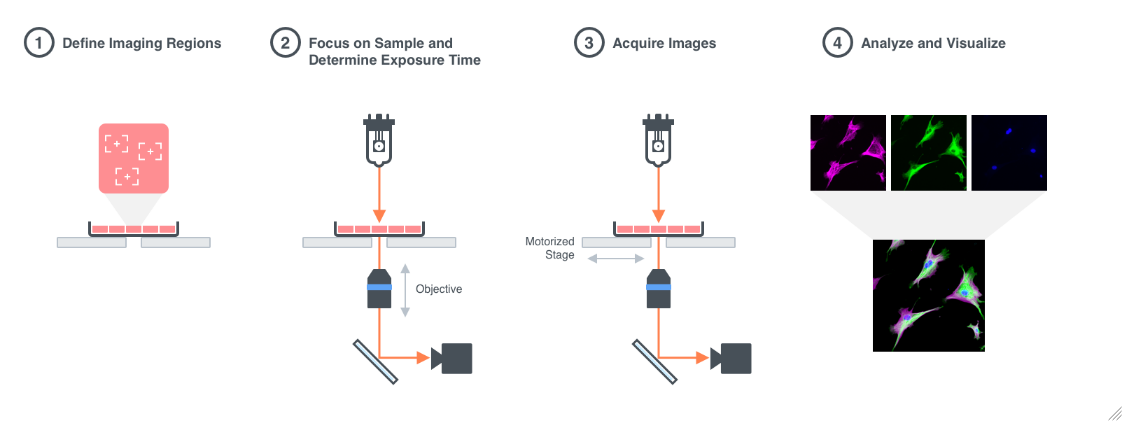
Figure 1.1: Imaging Process: 1. A sample is placed on the stage of a microscope and the regions to be imaged are defined. Optionally, a Time-series and/or Z-series imaging can be set up. 2. The objective lens is focused on the sample and the camera exposure time is determined. 3. Images are acquired using a camera connected to the microscope. 4. Images are analyzed.
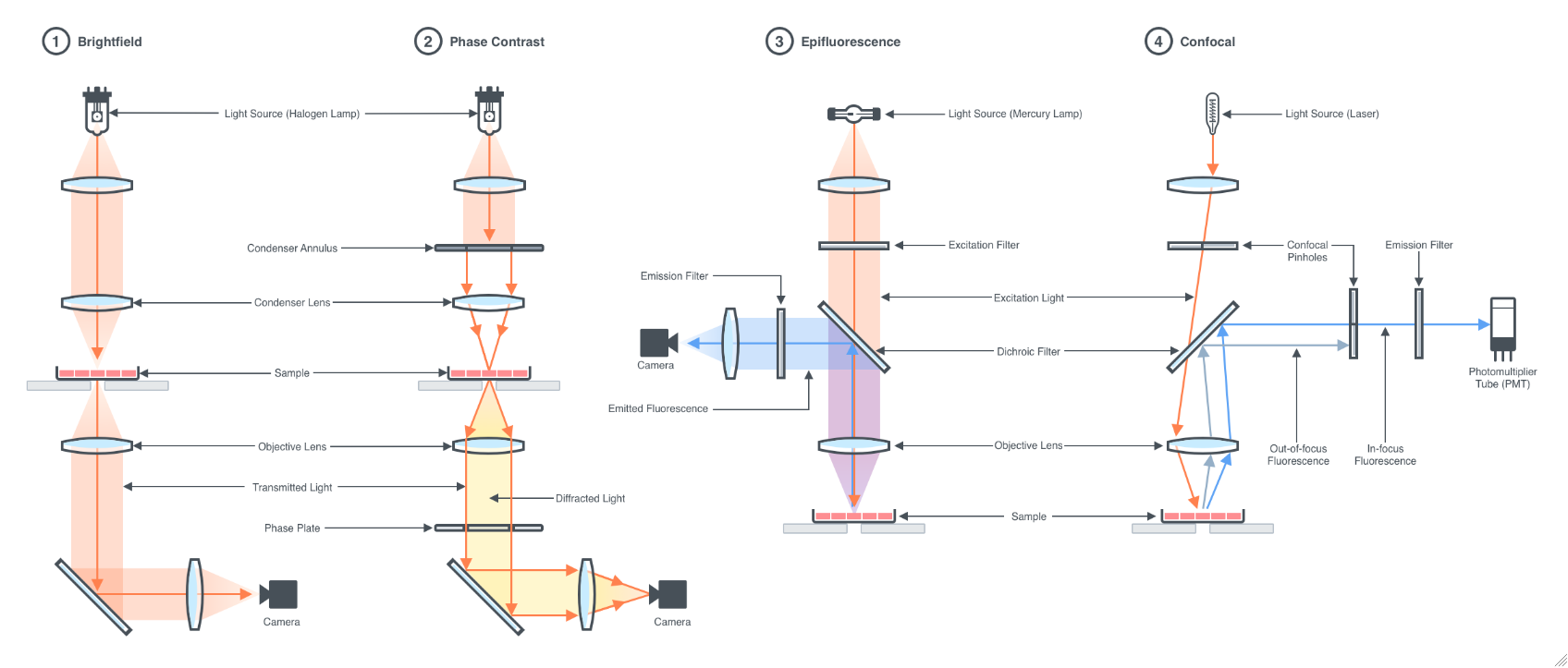
Figure 1.2: Microscopy Techniques: 1. Bright-field is the simplest type of microscopy technique that uses white light to illuminate the sample, resulting in an image in which the specimen is darker than the surrounding areas that are devoid of it. Bright-field microscopy is useful for imaging samples stained with dyes or samples with intrinsic colors. 2. Phase-contrast microscopy increases the contrast between the sample and its background, allowing it to produce highly detailed images from living cells and transparent biological samples. 3. Epifluorescence microscopy uses light at a specific wavelength range to excite a fluorophore of interest in the sample and capture the resulting emitted fluorescence to generate an image. 4. Confocal microscopy employs a similar principle as Epifluorescence to illuminate the sample and capture the emitted fluorescence along with pinholes in the light path to block out-of-focus light in order to increase optical resolution. Confocal microscopy is often used to image thick samples or to clearly distinguish structures that vary in height along the z-axis.
Instrumentation
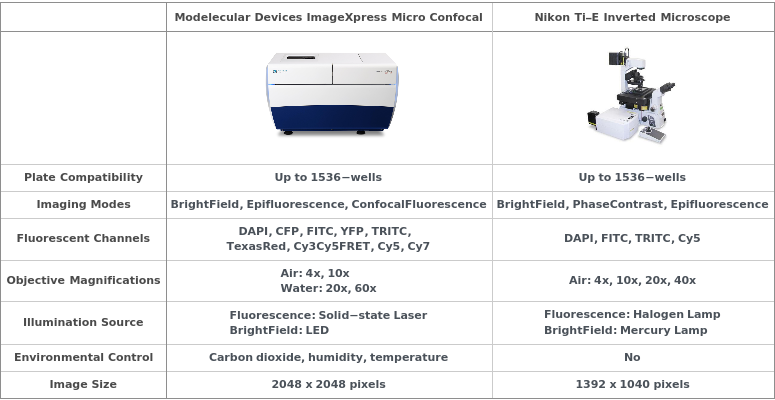
Molecular Devices ImageXpress Micro Confocal

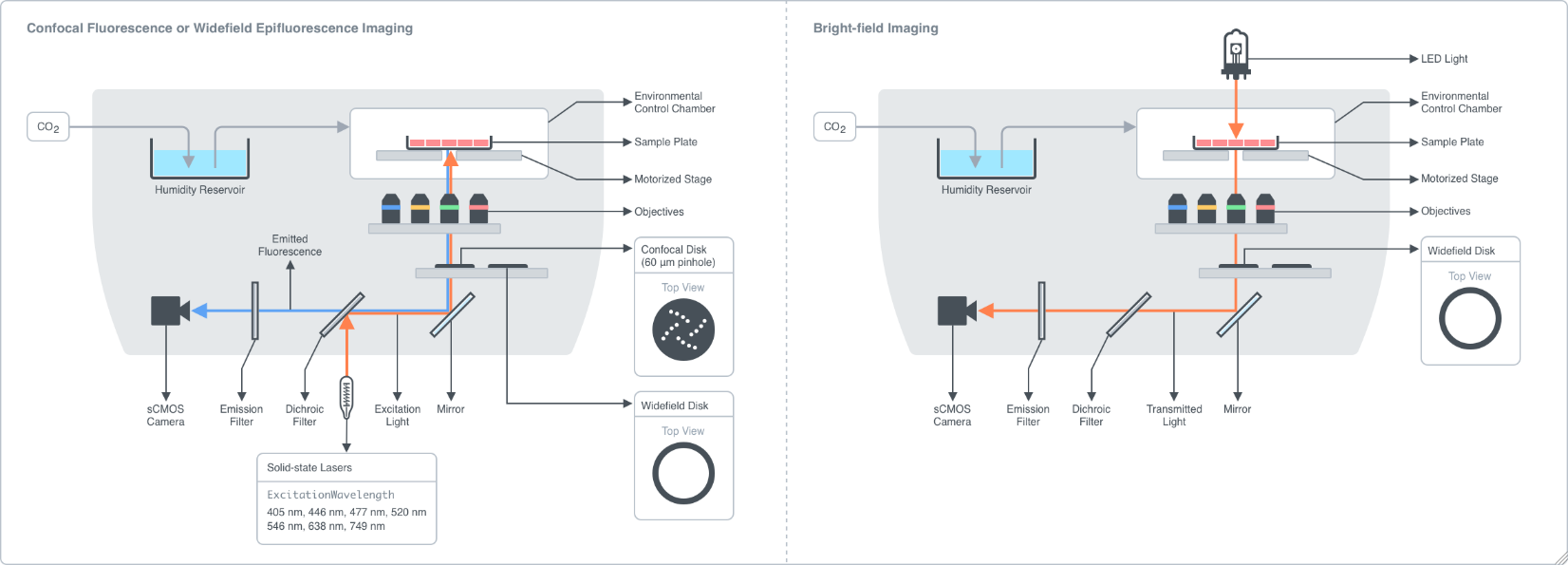
Figure 2.1.1: The high-content imager is a fluorescence microscope capable of automating imaging of a large number of samples. Samples are placed onto a motorized stage that allows accurate positioning and alignment of each sample with the objective lenses. The imager is equipped with an environmental control chamber to maintain optimal temperature, humidity, and carbon dioxide levels for extended time-series imaging of live cells. A solid-state laser system and a LED light source enable fluorescence imaging and bright-field imaging of samples. Illumination light from the laser is directed through a dichroic filter which selectively reflects light at a certain wavelength range while transmits light at a different wavelength range. The reflected light passes through the objective lens and to the sample where it excites fluorophores of interest. The fluorescence emission from the sample is directed back into the light path, transmitted through the dichroic filter, and an emission filter which allows light within a certain wavelength range to be captured by a scientific CMOS camera (sCMOS). Different combinations of the excitation wavelengths, emission filters, and dichroic filters may be specified to image a wide range of fluorophores. The high-content imager can also operate in confocal mode for imaging thicker specimens, such as tissue sections. In confocal mode, a spinning disk consisting of an array of 60-μm pinholes is placed into the light path just before the objective lens to block out-of-focus signal from being captured. As a result, the images can be acquired with a better optical resolution.
Ti-E Inverted Microscope

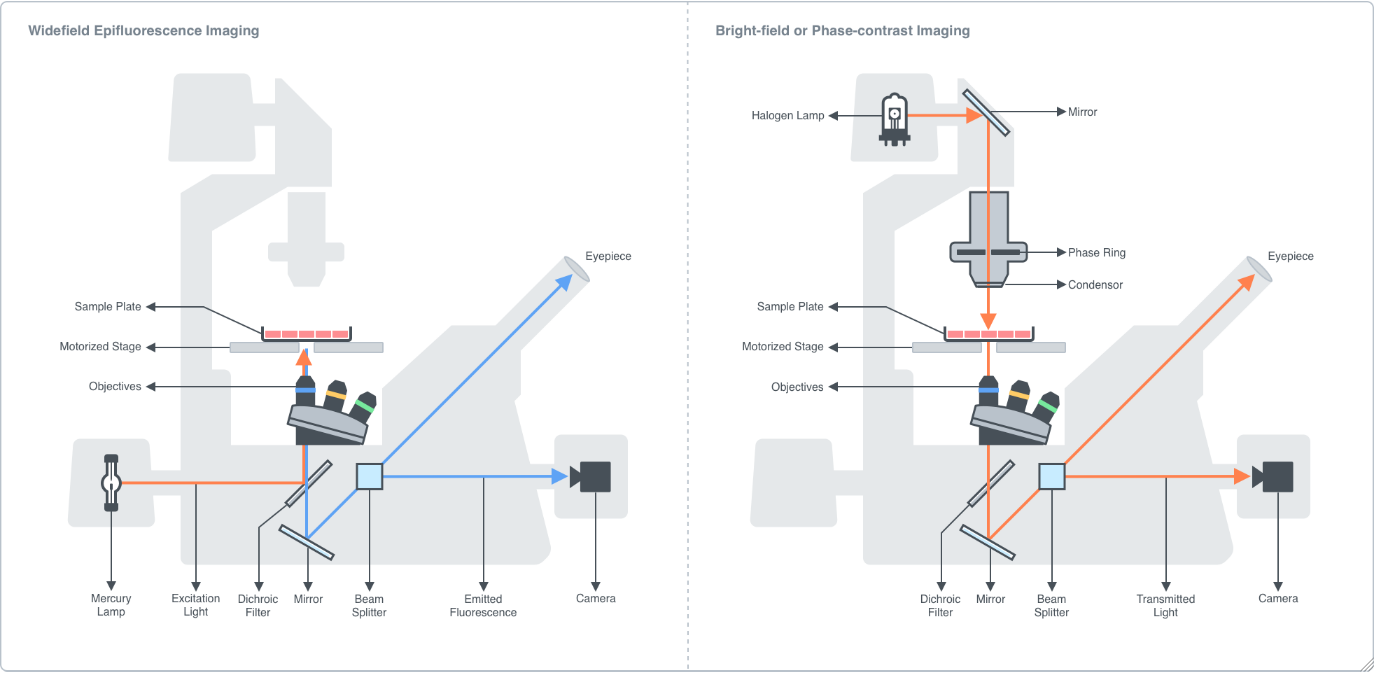
Figure 2.2.1: Instrument diagram of an inverted epifluorescence widefield microscope. A sample is placed onto a motorized stage and brought into focus by adjusting the coarse and fine focus adjustment knobs which move the objective turret in the Z-direction. The motorized stage enables translocation of the sample in the XY-direction. In epifluorescence mode, white light from a mercury lamp is passed through an excitation filter which only allows light within a certain wavelength range through. The beam is then directed to a dichroic filter which selectively reflects light in the excitation wavelength range and transmits light in the emission wavelength range. The reflected excitation light is directed through the objective lens and to the sample where it excites a fluorophore. The fluorescence emission of the fluorophore is directed back into the objective, through the dichroic mirror, and through the emission filter, which only allows light in a specified wavelength range to pass through. The emission fluorescence is then directed into a beam splitter which directs the signal to either the camera or the eyepiece. The epifluorescence microscope is equipped with a range of fluorescence filter cube sets to image at 4 different imaging channels commonly used for biological samples: DAPI (blue), FITC (green), TRITC (red), and Cy5 (far red).
In bright-field or phase-contrast mode, the sample is illuminated by a halogen lamp. Phase-contrast microscopy enhances the contrast between the sample and background, allowing for more detailed imaging of clear biological samples. In phase-contrast mode, an annulus is placed in the light path to generate a ring-shaped beam that when illuminates the sample, some light is scattered, and some is transmitted (background light). The scattered light is then phase-shifted by the phase plate located within the body of the objective lens, which destructively interferes with the background light to reduce the light intensity in regions where a specimen is, resulting in an image where the specimen is darker than the surrounding areas that are devoid of it.
AcquireImage
Mode
The type of microscopy technique used to acquire an image of a sample. BrightField is the simplest type of microscopy technique that uses white light to illuminate the sample, resulting in an image in which the specimen is darker than the surrounding areas that are devoid of it. Bright-field microscopy is useful for imaging samples stained with dyes or samples with intrinsic colors. PhaseContrast microscopy increases the contrast between the sample and its background, allowing it to produce highly detailed images from living cells and transparent biological samples. Epifluorescence microscopy uses light at a specific wavelength range to excite a fluorophore of interest in the sample and capture the resulting emitted fluorescence to generate an image. ConfocalFluorescence microscopy employs a similar principle as Epifluorescence to illuminate the sample and capture the emitted fluorescence along with pinholes in the light path to block out-of-focus light in order to increase optical resolution. Confocal microscopy is often used to image thick samples or to clearly distinguish structures that vary in height along the z-axis.
General
Mode
The type of microscopy technique used to acquire an image of a sample. BrightField is the simplest type of microscopy technique that uses white light to illuminate the sample, resulting in an image in which the specimen is darker than the surrounding areas that are devoid of it. Bright-field microscopy is useful for imaging samples stained with dyes or samples with intrinsic colors. PhaseContrast microscopy increases the contrast between the sample and its background, allowing it to produce highly detailed images from living cells and transparent biological samples. The contrast is enhanced such that the boundaries of the sample and its structures appear much darker than the surrounding medium. Epifluorescence microscopy uses light at a specific wavelength range to excite a fluorophore of interest in the sample and capture the resulting emitted fluorescence to generate an image. ConfocalFluorescence microscopy employs a similar principle as Epifluorescence to illuminate the sample and capture the emitted fluorescence along with pinholes in the light path to block out-of-focus light in order to increase optical resolution. Confocal microscopy is often used to image thick samples or to clearly distinguish structures that vary in height along the z-axis.
Default Calculation: Automatically set to Epifluorescence if any fluorophore is present in DetectionLabels Field of the sample's identity model. Otherwise, set to BrightField if DetectionLabels is an empty list.
DetectionLabels
Indicates the tags, including fluorescent or non-fluorescent chemical compounds or proteins, attached to the sample that will be imaged.
Default Calculation: Automatically set to an object or a list of objects present in the DetectionLabels Field of the sample's identity model that does not exist in other AcquireImage primitives.
Pattern Description: An object of type or subtype Model[Molecule] or list of one or more an object of type or subtype Model[Molecule] entries or Null.
Programmatic Pattern: ((ObjectP[Model[Molecule]] | {ObjectP[Model[Molecule]]..}) | Automatic) | Null
ImagingChannel
Indicates the imaging channel pre-defined by the instrument that should be used to acquire images from the sample.
Default Calculation: If none of the options in Fluorescence Imaging or BrightField and PhaseContrast Imaging is specified, automatically set to the instrument's imaging channel with ExcitationWavelength and EmissionWavelength capable if illuminating and detecting signal from DetectionLabels.
Pattern Description: DAPI, CFP, FITC, YFP, TRITC, TexasRed, Cy3Cy5FRET, Cy3, Cy5, Cy7, CustomChannel, or TransmittedLight or Null.
Image Acquisition
FocalHeight
The distance between the top of the objective lens and the bottom of the sample when the imaging plane is in focus. If set to Autofocus, the microscope will obtain a small stack of images along the z-axis of the sample and determine the best focal plane based on the image in the stack that shows the highest contrast. FocalHeight is then calculated from the location of the best focal plane on the z-axis. If set to Manual, the FocalHeight will be adjutsted manually.
Default Calculation: Automatically set to Autofocus if the selected instrument supports autofocusing. Otherwise, set to Manual.
ExposureTime
The length of time that the camera collects the signal from the sample. The longer the exposure time, the more photons the detector can collect, resulting in a brighter image. Selecting AutoExpose will prevent the pixels from becoming saturated by allowing the microscope software to determine the exposure time such that the brightest pixel is 75% of the maximum gray level that the camera can obtain.
TargetMaxIntensity
Specifies the intensity that the instrument should attempt to attain for the brightest pixel in the image to be acquired.
Time Lapse Imaging
TimelapseImageCollection
Specifies the timepoint(s) at which the sample is imaged when acquiring timelapse images. All allows acquisition of an image at every timepoint defined in ExperimentImageCells option. StartOnly acquires an image at the first timepoint only. StartAndEnd acquires an image at the first and the last timepoints. When specified as an integer, an image will be acquired at every Nth timepoints, beginning with the first timepoint.
Default Calculation: Automatically set to All if Timelapse option of ExperimentImageCells is set to True.
Z-Stack Imaging
ZStackImageCollection
Default Calculation: Automatically set to True if ZStack option of ExperimentImageCells is set to True.
Fluorescence Imaging
ExcitationWavelength
The wavelength of excitation light used to illuminate the sample when imaging with ConfocalFluorescence or Epifluorescence Mode.
EmissionWavelength
Default Calculation: Automatically set to the wavelength closest to the fluorescence emission wavelength of the DetectionLabels.
ExcitationPower
The percent of maximum intensity of the light source that should be used to illuminate the sample. Higher intensity will excite more fluorescent molecules in the sample, resulting in more signal being produced, but will also increase the chance of bleaching the fluorescent molecules.
Default Calculation: Automatically set to 20% if a high content imager is selected as instrument. Otherwise, set to 100% if Mode is ConfocalFluorescence or Epifluorescence
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
DichroicFilterWavelength
Specifies the wavelength that should be passed by the filter to illuminate the sample and excite the DetectionLabels.
Default Calculation: Automatically set to wavelength closest to the fluorescence excitation wavelength of the DetectionLabels.
ImageCorrection
The correction step(s) that will be automatically applied to the image. BackgroundCorrection removes stray light that is unrelated to light that reaches the sample. ShadingCorrection mitigates the uneven illumination of the sample that is visible around the edges of the image.
Default Calculation: Automatically apply both BackgroundCorrection AND ShadingCorrection to the image if the high content imager is selected as Instrument and Mode is either ConfocalFluorescence or Epifluorescence. Otherwise, set to ShadingCorrection when Mode is BrightField.
Pattern Description: BackgroundCorrection, ShadingCorrection, or BackgroundAndShadingCorrection or Null.
ImageDeconvolution
Indicates if a deconvolution algorithm should be used to enhance contrast, improve image resolution, and sharpen the image.
ImageDeconvolutionKFactor
Specifies the factor used by the Wiener Filter in the deconvolution algorithm to determine image sharpness. Lower values increase sharpness and higher values reduce noise.
BrightField and PhaseContrast Imaging
TransmittedLightPower
The percent of maximum intensity of the transmitted light that should be used to illuminate the sample. This option will set the percent maximum of the voltage applied to the light source, with higher percentages indicating higher intensities.
Default Calculation: If Mode is BrightField or PhaseConstrast, automatically set to 20% for a high content inager, or 100% for {Model[Instrument, Microscope, Ti-E Inverted Microscope], Object[Instrument, Microscope, Epifluorescence inverted microscope]}.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
TransmittedLightColorCorrection
Indicates if a neutral color balance filter will be placed into the transmitted light path to correct the color temperature during BrightField and PhaseContrast imaging.
Experiment Options
General
MicroscopeOrientation
The location of the objective lens relative to the sample on the microscope stage. Inverted refers to having the objective lens below the sample. Only Inverted microscopes are currently available at ECL. This option can only be set if Preparation -> Manual.
Default Calculation: Automatically set to Orientation of Instrument. Otherwise, set to Inverted if Preparation -> Robotic.
ReCalibrateMicroscope
Indicates if the optical components of the microscope should be adjusted before imaging the sample. This option can only be set to True if Preparation -> Manual.
MicroscopeCalibration
A calibration object that specifies a set of parameters used to adjust optical components of the microscope. This option can only be set if Preparation -> Manual.
Default Calculation: If CalibrateMicroscope is True, automatically set to the most recent Object[Maintenance,CalibrateMicroscope] performed on the selected instrument.
SampleLabel
The label of the cell samples that are being imaged, which is used for identification elsewhere in cell preparation.
SampleContainerLabel
The label of the cell sample's container that are being imaged, which is used for identification elsewhere in cell preparation.
Instrument
Default Calculation: Automatically set to the high content imager instrument model if PhaseContrast is not included as Mode in AcquireImage primitives specified in the Images option.
Pattern Description: An object of type or subtype Model[Instrument, Microscope] or Object[Instrument, Microscope]
Programmatic Pattern: ObjectP[{Model[Instrument, Microscope], Object[Instrument, Microscope]}] | Automatic
Preparation
Indicates if this unit operation is carried out primarily robotically or manually. Manual unit operations are executed by a laboratory operator and robotic unit operations are executed by a liquid handling work cell.
Image Acquisition
ContainerOrientation
For each sample pool, the orientation of the sample container when placed on the microscope stage for imaging. RightSideUp refers to placing the sample container on the stage such that the cover is on the top (plates: lids on top, microscope slides: coverslip facing up). UpsideDown refers to inverting the sample container on the stage such that the cover faces down (plates: lids facing down, microscope slides: coverslip facing down).
Default Calculation: Automatically set RightSideUp if Preparation -> Robotic. Otherwise set to RightSideUp if sample's container is a plate or UpsideDown if sample's container is a slide.
CoverslipThickness
For each sample pool, the thickness of the coverslip, which is used to determine the position of the CorrectionCollar on high magnification objective lenses to correct for optical aberrations. This option can only be set if Preparation -> Manual.
Default Calculation: Automatically set to the thickness of the coverslip specified in the container model of the sample.
Pattern Description: Greater than or equal to 0 millimeters and less than or equal to 2 millimeters or Null.
EquilibrationTime
For each sample pool, specifies the amount of time for which the samples are placed on the stage of the microscope before the first image is acquired.
Default Calculation: Automatically set to 5 minutes for sample's that contains Model[Cell] with CellType -> Suspension.
PlateBottomThickness
For each sample pool, the thickness of the well bottom of the sample's container, which is used to determine the position of the CorrectionCollar on high magnification objective lenses to correct for optical aberrations.
Default Calculation: Automatically set to the well bottom thickness specified in the container model of the sample.
Pattern Description: Greater than or equal to 0 millimeters and less than or equal to 5 millimeters or Null.
PixelBinning
For each sample pool, the "n x n" grid of pixels whose intensity values should be combined into a single pixel. Higher binning values result in higher overall signal-to-noise ratios but lower pixel resolutions.
Images
For each sample pool, a list of acquisition parameters used to image a sample. Each list of acquisition parameters corresponds to a single output image acquired from the input sample.
Default Calculation: Automatically generates a unique image acquisition for each of the fluorophores present in the DetectionLabels Field of the sample's identity model. The value of acquisition parameters are determined from the Mode and the DetectionLabels of the sample's identity model.
Pattern Description: A primitive with head AcquireImage or list of one or more a primitive with head AcquireImage entries.
AdjustmentSample
For each sample pool, specifies the sample should be used to adjust the ExposureTime and FocalHeight in each imaging session. In each sample pool, only one sample is allowed to be used as AdjustmentSample. If set to All, the ExposureTime and FocalHeight will be adjusted for each sample individually.
Environmental Controls
Temperature
Pattern Description: Ambient or greater than or equal to 25 degrees Celsius and less than or equal to 40 degrees Celsius.
CarbonDioxide
CarbonDioxidePercentage
Optics
ObjectiveMagnification
For each sample pool, the magnification power defined by the ratio between the dimensions of the image and the sample. Low magnification power (4X and 10X) is recommended for acquiring an overview image of a sample. High magnification power (20X, 40X, 60X) is recommended for imaging detailed structure of a large tissue sample or intracellular structures.
Default Calculation: Automatically set to 10 if available on the selected instrument. Otherwise, set to the lowest magnification supported by the selected instrument.
Sampling
SamplingPattern
For each sample pool, the pattern of images that will be acquired from the samples. SinglePoint: acquires an image at the center of each well or sample. Grid: acquires multiple images along specified rows and columns. Coordinates: acquires image(s) at requested coordinates in each well or on a microscopic slide. Adaptive: uses an algorithm to calculate the number of cells in each field of view to increase the chances of acquiring valid data, until the indicated number of cells specified in AdaptiveNumberOfCells is obtained.
Sampling Regions
SamplingNumberOfRows
For each sample pool, the number of rows which will be acquired for each sample if the Grid SamplingPattern is selected. If Adaptive SamplingPattern is selected, SamplingNumberOfRows specifies maximum number of rows to be imaged if AdaptiveNumberOfCells cannot be reached.
Default Calculation: Automatically set to maximum number of rows that can fit within the sample's container if Grid or Adaptive is selected as SamplingPattern.
SamplingNumberOfColumns
For each sample pool, the number of columns which will be acquired for each sample if the Grid SamplingPattern is selected. If Adaptive SamplingPattern is selected, SamplingNumberOfColumns specifies maximum number of columns to be imaged if AdaptiveNumberOfCells cannot be reached.
Default Calculation: Automatically set to maximum number of columns that can fit within the sample's container if Grid or Adaptive is selected as SamplingPattern.
SamplingRowSpacing
For each sample pool, the distance between each row of images to be acquired. Negative distances indicate overlapping regions between adjacent rows. Overlapping regions between rows can also be specified as percentage if desired.
Default Calculation: Automatically set to 0 Micrometer if Grid or Adaptive is selected as SamplingPattern.
Programmatic Pattern: ((RangeP[-85*Millimeter, 85*Millimeter] | RangeP[0*Percent, 100*Percent]) | Automatic) | Null
SamplingColumnSpacing
For each sample pool, the distance between each column of images to be acquired. Negative values indicate overlapping regions between adjacent columns. Overlapping regions between columns can also be specified as percentage if desired.
Default Calculation: Automatically set to 0 Micrometer if Grid or Adaptive is selected as SamplingPattern.
Programmatic Pattern: ((RangeP[-127*Millimeter, 127*Millimeter] | RangeP[0*Percent, 100*Percent]) | Automatic) | Null
SamplingCoordinates
For each sample pool, specifies the positions at which images are acquired. The coordinates are referenced to the center of each well if the sample's container is a plate or center of a microscopic slide, which has coordinates of (0,0).
Default Calculation: If the SamplingPattern is set to Coordinates, SamplingCoordinates will be set to randomly acquire images from 3 different sites in each well or on a microscopic slide.
Programmatic Pattern: ({{RangeP[-127*Millimeter, 127*Millimeter], RangeP[-85*Millimeter, 85*Millimeter]}..} | Automatic) | Null
Adaptive Sampling
AdaptiveExcitationWaveLength
For each sample pool, specifies the excitation wavelength of the light source that will be used to determine the number of cells in each field of view for the Adaptive SamplingPattern.
Default Calculation: Automatically set to the ExcitationWavelength indicated in the first AcquireImage primitive if Adaptive is selected as SamplingPattern.
AdaptiveNumberOfCells
For each sample pool, specifies the minimum cell count per well that the instrument will work to satisfy by acquiring images from multiple regions before moving to the next sample.
AdaptiveMinNumberOfImages
For each sample pool, the minimum number of regions that must be acquired when using the Adaptive SamplingPattern, even if the specified cell count (AdaptiveNumberOfCells) is already reached.
Default Calculation: Automatically set to 1 imaging site if Adaptive is selected as SamplingPattern.
AdaptiveCellWidth
For each sample pool, specifies the expected range of cell size in the sample. The instrument uses this range to determine which features in the image will be counted as cells.
Pattern Description: A span from anything greater than or equal to 0 micrometers and less than or equal to 100 micrometers to anything greater than or equal to 0 micrometers and less than or equal to 100 micrometers or Null.
Programmatic Pattern: (RangeP[0*Micrometer, 100*Micrometer] ;; RangeP[0*Micrometer, 100*Micrometer] | Automatic) | Null
AdaptiveIntensityThreshold
For each sample pool, the intensity above local background value that a putative cell needs to have in order to be counted. The intensity above local background value is calculated by subtracting the gray value of the surrounding background from the gray level value of a putative cell. Any feature in an image is considered to be a putative cell if its size falls within the specified cell width (AdaptiveCellWidth).
Time Lapse Imaging
Timelapse
TimelapseInterval
For each sample pool, the amount of time between the start of an acquisition at one time point and the start of an acquisition at the next time point.
TimelapseDuration
For each sample pool, the total amount of time that the Timelapse images will be acquired for the sample.
NumberOfTimepoints
For each sample pool, the number of images that will be acquired from the sample in during the course of Timelapse imaging.
Default Calculation: Automatically set to the maximum number of images that can be acquired in the course of TimelapseDuration with the amount of time between each image specified by TimelapseInterval.
ContinuousTimelapseImaging
For each sample pool, indicates if images from multiple time points should be acquired from the samples in a single imaging session.
Default Calculation: Automatically set to True if Timelapse is True and TimelapseInterval is less than 1 Hour.
TimelapseAcquisitionOrder
For each sample pool, determines the order in which the time-series images are acquired with respect to the sample's location in the plate. Parallel acquires images from every selected well in the plate at each time point before moving on to the next time point. Serial acquires images for all time points in one well before moving on to the next well. RowByRow acquires images at each time point in each row of wells before moving on to the next row. ColumnByColumn acquires images at each time point in each column of wells before moving on to the next column.
Z-Stack Imaging
ZStack
For each sample pool, indicates if a series of images at multiple z-axis positions will be acquired for the sample.
ZStepSize
Default Calculation: Automatically set to the Nyquist sampling distance of the objective by calculating from the following equation: ExcitationWavelength/2n(1-cos(α)). ExcitationWavelength is set to default value of 550 nm. α = arcsin(NA/n) where n is refractive index of the objective's immersion medium. NA is numerical aperture of the objective.
NumberOfZSteps
Default Calculation: Automatically set to 10 if ZStack is True and no other Z-stack options are selected, or calculated from the ZStackSpan and ZStepSize options if specified.
ZStackSpan
For each sample pool, the range of Z-heights that the microscope will acquire images from in a Z-Stack. Negative values indicate planes that are below the sample's focal plane (as defined in FocalHeight) whereas positive values indicate planes that are above the sample's focal plane.
Default Calculation: If ZStack is True, automatically calculated from the NumberOfZsteps and ZStepSize.
Pattern Description: A span from anything greater than or equal to 0 micrometers and less than or equal to 10000 micrometers to anything greater than or equal to -10000 micrometers and less than or equal to 0 micrometers or Null.
Programmatic Pattern: (RangeP[-10000*Micrometer, 0*Micrometer] ;; RangeP[0*Micrometer, 10000*Micrometer] | Automatic) | Null
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
PreparatoryPrimitives
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSampleManipulation.
Pattern Description: List of one or more a primitive with head Define, Transfer, Mix, Aliquot, Consolidation, FillToVolume, Incubate, Filter, Wait, Centrifuge, or Resuspend entries or Null.
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Object[Instrument, Roller], Model[Instrument, OverheadStirrer], Object[Instrument, OverheadStirrer], Model[Instrument, Vortex], Object[Instrument, Vortex], Model[Instrument, Shaker], Object[Instrument, Shaker], Model[Instrument, BottleRoller], Object[Instrument, BottleRoller], Model[Instrument, Roller], Object[Instrument, Roller], Model[Instrument, Sonicator], Object[Instrument, Sonicator], Model[Instrument, HeatBlock], or Object[Instrument, HeatBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Roller], Object[Instrument, Roller], Model[Instrument, OverheadStirrer], Object[Instrument, OverheadStirrer], Model[Instrument, Vortex], Object[Instrument, Vortex], Model[Instrument, Shaker], Object[Instrument, Shaker], Model[Instrument, BottleRoller], Object[Instrument, BottleRoller], Model[Instrument, Roller], Object[Instrument, Roller], Model[Instrument, Sonicator], Object[Instrument, Sonicator], Model[Instrument, HeatBlock], Object[Instrument, HeatBlock]}] | Automatic) | Null
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, Silica, or HLB or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.1 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotSampleLabel
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentrationFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Warnings and Errors
Messages (73)
CarbonDioxideOptionsMismatch (1)
CO2IncompatibleMicroscope (1)
FirstWavelengthNotUsedForAdaptive (1)
ImageCellsAdaptiveCellWidthNotAllowed (1)
ImageCellsAdaptiveCellWidthNotSpecified (1)
ImageCellsAdaptiveExcitationWaveLengthNotAllowed (1)
ImageCellsAdaptiveIntensityThresholdNotAllowed (1)
ImageCellsAdaptiveIntensityThresholdNotSpecified (1)
ImageCellsAdaptiveMinNumberOfImagesNotAllowed (1)
ImageCellsAdaptiveMinNumberOfImagesNotSpecified (1)
ImageCellsAdaptiveNumberOfCellsNotAllowed (1)
ImageCellsAdaptiveNumberOfCellsNotSpecified (1)
ImageCellsCannotDetermineImagingSites (1)
ImageCellsContinuousTimelapseImagingNotAllowed (1)
ImageCellsGridDefinedAsSinglePoint (1)
ImageCellsIncompatibleContainer (1)
ImageCellsIncompatibleContainerThickness (1)
ImageCellsInstrumentModelNotFound (1)
ImageCellsInvalidAcquireImagePrimitive (1)
ImageCellsInvalidAdaptiveExcitationWaveLength (1)
ImageCellsInvalidAdaptiveIntensityThreshold (1)
ImageCellsInvalidAdjustmentSample (1)
ImageCellsInvalidContinuousTimelapseImaging (1)
ImageCellsInvalidCoverslipThickness (1)
ImageCellsInvalidGridDefinition (1)
ImageCellsInvalidObjectiveMagnification (1)
ImageCellsInvalidPlateBottomThickness (1)
ImageCellsInvalidSamplingCoordinates (1)
ImageCellsInvalidTimelapseDefinition (1)
ImageCellsInvalidZStackDefinition (1)
ImageCellsInvalidZStackSpan (1)
ImageCellsInvalidZStepSize (1)
ImageCellsInvalidZStepSizeNumberOfZSteps (1)
ImageCellsMismatchedContainers (1)
ImageCellsMultipleContainersForAdjustmentSample (1)
ImageCellsNegativePlateBottomThickness (1)
ImageCellsNumberOfTimepointsNotAllowed (1)
ImageCellsNumberOfZStepsNotAllowed (1)
ImageCellsSamplingColumnSpacingMismatch (1)
ImageCellsSamplingColumnSpacingNotAllowed (1)
ImageCellsSamplingColumnSpacingNotSpecified (1)
ImageCellsSamplingCoordinatesNotAllowed (1)
ImageCellsSamplingCoordinatesNotSpecified (1)
ImageCellsSamplingNumberOfColumnsNotAllowed (1)
ImageCellsSamplingNumberOfRowsNotAllowed (1)
ImageCellsSamplingRowSpacingMismatch (1)
ImageCellsSamplingRowSpacingNotAllowed (1)
ImageCellsSamplingRowSpacingNotSpecified (1)
ImageCellsTimelapseAcquisitionOrderNotAllowed (1)
ImageCellsTimelapseDurationNotAllowed (1)
ImageCellsTimelapseIntervalNotAllowed (1)
ImageCellsTooManyAdaptiveImagingSites (1)
ImageCellsUnsupportedSamplingPattern (1)
ImageCellsUnsupportedTimelapseImaging (1)
ImageCellsUnsupportedZStackImaging (1)
InvalidMicroscopeCalibration (1)
MicroscopeCalibrationMismatch (1)
MicroscopeCalibrationNotAllowed (1)
MicroscopeCalibrationNotFound (1)
MicroscopeOrientationMismatch (1)
MismatchedCoverslipThickness (1)
Possible Issues
Photobleaching
Fluorophore molecules may can lose the ability to fluoresce after absorbing photons from illumination light after multiple rounds of imaging. It is highly recommendeded that minimal ExposureTime and ExcitationPower are used to prevent photobleaching.

Well-to-well Crosstalk
Imaging samples in plates with clear walls may increase the chance of acquiring unwanted fluorescence from adjacent wells. It is highly recommendeded that plates with opaque black walls are used.

Background Fluorescence
Imaging samples in plates with glass well bottom and opaque black walls can significantly reduce background fluorescence generated by illuminating polystyrene well bottom and non-opaque materials.

Last modified on Thu 3 Mar 2022 10:20:42