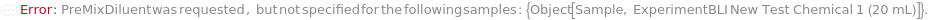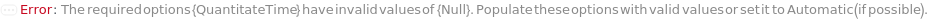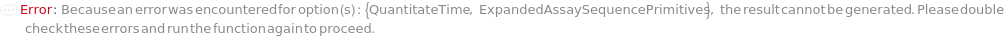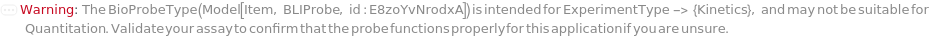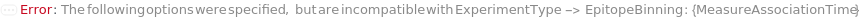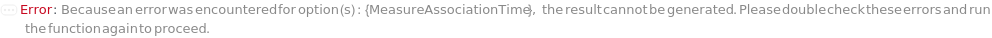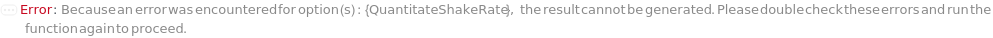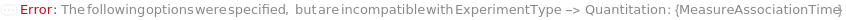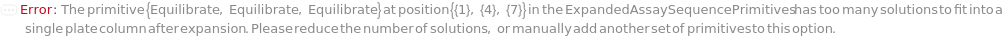ExperimentBioLayerInterferometry
ExperimentBioLayerInterferometry[Samples]⟹Protocol
generates a Protocol object for performing Bio-Layer Interferometry (BLI) on the provided Samples.
Biolayer interferometry is a technique used to quantify the magnitude and kinetics of an interaction between a surface immobilized species and a solution phase analyte. Binding of the analyte is monitored by the change in thickness of the bound bio-layer on the tip of a fiber-optic probe, which is determined by the white-light interference pattern of a reference channel and the bio-layer/solution interface. Probe tips can be functionalized via immersion in a solution of the target immobilized species and regenerated to return either a loading ready or measurement ready surface. After appropriate treatment, the probes are dipped into a sample solution to quantify the analyte concentration or to determine the association and dissociation kinetics. Other common experiments include epitope binning and assay optimization. Biolayer interferometry is useful for studying a variety of interactions involving proteins, oligomers, small molecules, fragments, and antibodies, and can be performed on crude samples such as cell lysate.
Experimental Principles
Figure 1.1: Procedural overview of a Biolayer Interferometry experiment. Step 1: The assay plate is loaded with buffers, loading solutions, standards, analytes, and other types of solutions required for the desired assay. Step 2: The probes are loaded into the probe rack position, where they may be immersed in equilibration solution prior to and during the course of the assay. Step 3: The multi-channel reader probes are immersed in an assay plate column to load the probe surface with an immobilized species (ActivateSurface, LoadSurface, Quench), measure the analyte binding properties (MeasureAssociation, MeasureDissocaition, Quantitate), or return the probe to measurement-ready condition via regeneration (Regenerate, Neutralize, Wash). Step 4: Any solutions derived from a given sample can be recovered from the AssayPlate.
Instrumentation
Octet Red96e
Figure 2.1: Fiber-optic probes are stored in the probe rack position, either dry or immersed in a buffer solution. An 8-channel manifold moves between the probe rack and assay plate positions to pick up probes and conduct measurements via immersion in wells of a given column in the assay plate. A plate cover can be used to cover columns of the assay plate not currently being measured by the 8-channel reader to prevent evaporation over the course of long assays. The white light input reflects off of the internal optical layer and the biolayer/solution interface, producing an interference pattern which is directly proportional to the biolayer thickness.
Experiment Options
General
Instrument
Pattern Description: An object of type or subtype Model[Instrument, BioLayerInterferometer] or Object[Instrument, BioLayerInterferometer]
Programmatic Pattern: ObjectP[{Model[Instrument, BioLayerInterferometer], Object[Instrument, BioLayerInterferometer]}]
ExperimentType
The objective of bio-layer interferometry experiment. Kinetics: Measure association and dissociation rates of the interaction between a solution phase species and a functionalized bio-probe surface. Quantitation: Quantify the amount of analyte in a solution by measuring the change in bio-layer thickness upon immersion of a functionalized bio-probe in a solution containing the target analyte. EpitopeBinning: Use competition experiments between two antibodies to identify groups of antibodies which bind a given antigen at the same epitope. AssayDevelopment: Optimize the bio-probe functionalization and individual assay steps by performing steps in a variety of solutions.
BioProbeType
Indicates the type of surface-functionalized fiber-optic probe to be used in this experiment. The probe surface functionalization may allow for direct binding to the analyte, or can be further functionalized with an immobilized secondary species, which would then interact with the solution phase analyte in a subsequent step.
NumberOfRepeats
The number of times that the assay will be repeated for each set of samples. All assay steps will be repeated with a new set of bio-probes unless a Regeneration option has been selected.
Pattern Description: Greater than or equal to 2 and less than or equal to 12 in increments of 1 or Null.
AcquisitionRate
Indicates the number of recorded data points per second. A lower acquisition rate (2 Hz) will average more scans per data point, leading to better signal-to-noise ratios. A higher rate (10 Hz) will generates more data points per second, and is best suited for fast binding events which cause rapid changes in bio-layer thickness. The default value of 5 Hz balances data density with signal-to-noise ratio reduction and is suitable for most experiments.
PlateCover
Indicates if a plate cover should be included. This is recommended for assays which are expected to run for more than 4 hours in order to prevent evaporation.
Temperature
Pattern Description: Ambient or greater than or equal to 15 degrees Celsius and less than or equal to 40 degrees Celsius.
RecoupSample
Indicates if the SampleIn used for bio-layer interferometry measurement will be transferred back into the container that they were in prior to the measurement.
SaveAssayPlate
Indicates if the Assay Plate should be saved and stored or discarded after the assay is completed. Note that if both SaveAssayPlate and RecoupSample are True, input samples will be transferred out of the plate and the plate will be stored separately.
DefaultBuffer
The primary buffer solution to be used as the default solution for baseline, equilibration, bio-probe rinsing, or dissociation steps. Note that all assay steps using this solution will occur in a single set of wells containing DefaultBuffer, with the exception of Dissociation steps. If multiple buffers are required, indicate the appropriate buffers in the assay specific options section, or by using the AssaySequencePrimitives.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ReuseSolution
Indicates groups of assay step types may share a set of wells if they are performed on an identical set of solutions. For example, selecting MeasureDissociation and MeasureBaseline will allow baseline and dissociation steps to occur steps in the same set of buffer wells. A single step, such as Wash, can also be selected to indicate that all Wash using identical solution should be performed in the same set of wells. This will increase the number of wells for available for sample measurement.
Default Value: {{MeasureBaseline, Wash, Equilibrate, MeasureDissociation, LoadSurface, ActivateSurface, Quench, Regenerate, Neutralize, Quantitate, MeasureAssociation}}
Pattern Description: List of one or more list of one or more Equilibrate, Wash, ActivateSurface, Quench, LoadSurface, Regenerate, Neutralize, MeasureBaseline, MeasureAssociation, MeasureDissociation, or Quantitate entries entries or Null.
Blank
The solution which does not contain the analyte to be use as a negative control. This solution can be used to provide a baseline for a given assay step, to account for non-specific binding of other solution species, or to verify the reproducibility of a given experiment.
Default Calculation: Blank will resolve to the same value as the DefaultBuffer when an experiment requires a negative control, and Null if it is not required for the assay.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Standard
The solution, typically containing a concentration of analyte, to be used as a positive control. This can be used to account for baseline drift and changes in the bio-probe properties.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Assay Preparation
ProbeRackEquilibration
Indicates if the bio-probes used in the assay are equilibrated in ProbeRackEquilibrationBuffer solution while stored in the bio-probe rack, which holds bio-probes before and during the experiment. All bio-probes used in this experiment will remain immersed in the ProbRackEquilibrationBuffer until they are required for use in the assay.
ProbeRackEquilibrationTime
The minimum amount of time that the bio-probes used in the assay are equilibrated in ProbeRackEquilibrationBuffer prior to use in the assay. All bio-probes used in this experiment will remain immersed in the ProbeEquilibrationBufferSolution until required for the assay.
Default Calculation: ProbeRackEquilibrationTime will default to 10 Minute if ProbeRackEquilibration is True.
ProbeRackEquilibrationBuffer
Default Calculation: ProbeRackEquilibrationBuffer defaults to DefaultBuffer when ProbeRackEquilibration is true.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
StartDelay
Dictates the amount of time that the assay plate is located in the instrument at the desired temperature prior to beginning the assay.
Default Calculation: StartDelay is set to 15 Minutes if the PlateTemperature is not Ambient. This allows the assay plate temperature to equilibrate.
StartDelayShake
Equilibrate
Indicates if an assay step should be added to the beginning of the assay in which the bio-probes are equilibrated in the assay plate.
EquilibrateTime
The amount of time that the bio-probes are immersed in EquilibrateBuffer as the first step in the assay.
EquilibrateBuffer
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
EquilibrateShakeRate
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
Post Experiment
StandardStorageCondition
Specifies non-default conditions under which the Standard should be stored after the protocol is completed. If left unset, the Standard will be stored according to the current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
LoadSolutionStorageCondition
Specifies non-default conditions under which the LoadSolution should be stored after the protocol is completed. If left unset, the LoadSolution will be stored according to the current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
QuantitationStandardStorageCondition
Specifies non-default conditions under which the QuantitationStandard should be stored after the protocol is completed. If left unset, the QuantitationStandard will be stored according to the current StorageCondition.
Default Calculation: If Standard is used as the QuantitationStandard, this will be set to StandardStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
QuantitationEnzymeSolutionStorageCondition
Specifies non-default conditions under which the QuantitationEnzymeSolution should be stored after the protocol is completed. If left unset, the QuantitationEnzymeSolution will be stored according to the current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
BinningAntigenStorageCondition
Specifies non-default conditions under which the BinningAntigen should be stored after the protocol is completed. If left unset, the BinningAntigen will be stored according to the current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
TestInteractionSolutionsStorageConditions
Specifies non-default conditions under which the TestInteractionSolutions should be stored after the protocol is completed. If left unset, the TestInteractionSolutions will be stored according to the current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
TestLoadingSolutionsStorageConditions
Specifies non-default conditions under which the TestLoadingSolutions should be stored after the protocol is completed. If left unset, the TestLoadingSolutions will be stored according to the current StorageCondition.
Pattern Description: List of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal entries or Null.
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Dilution Preparation
DilutionMixVolume
The volume that is pipetted out and in of a given dilution to ensure homogeneous composition. This option also applies to PreMixSolution.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 2000 microliters or Null.
DilutionNumberOfMixes
The number of pipette out and in cycles that is used to mix each dilution. This option also applies to PreMixSolution.
Pattern Description: Greater than or equal to 0 and less than or equal to 20 in increments of 1 or Null.
DilutionMixRate
The rate at which the DilutionMixVolume is pipetted when mixing to ensure homogeneity. This option also applies to PreMixSolution.
Pattern Description: Greater than or equal to 0.5 microliters per second and less than or equal to 250 microliters per second or Null.
Probe Regeneration
RegenerationType
Indicates the steps to be included when returning the bio-probe surface to a measurement-ready condition. Regenerate: Adds a step where the bio-probe is immersed in RegenerationSolution. Neutralize: The bio-probe is immersed in NeutralizationSolution after regeneration to neutralize the probe surface. Wash: Adds a washing step after Regeneration or Neutralization (if selected) in WashSolution to remove any residual solution. PreCondition: Performs the requested regeneration cycle prior to the first sample measurement to ensure all measurements are performed with identical assay steps.
Pattern Description: A selection of one or more of Regenerate, Neutralize, Wash, or PreCondition or None or Null.
Programmatic Pattern: (None | DuplicateFreeListableP[Regenerate | Neutralize | Wash | PreCondition]) | Null
RegenerationSolution
The solution used to remove residual analyte from the bio-probe surface. Regeneration with the appropriate solution will allow for reuse of a set of bio-probes for multiple measurements.
Default Calculation: RegenerationSolution defaults to Model[Sample, StockSolution, "2 M HCl"] if Regenerate is selected in RegenerationType
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
RegenerationCycles
Indicates the number of times bio-probe will be subjected to the selected Regenerate and Neutralize steps (prior to a Wash step, if selected).
Default Calculation: RegenerationCycles will be set to 3 if RegenerationParameters is not None. This will generally provide a good balance between complete bio-probe regeneration and experiment time.
Pattern Description: Greater than or equal to 0 and less than or equal to 10 in increments of 1 or Null.
RegenerationTime
Default Calculation: RegenerationTime will be set to 5 Second if regeneration steps are included in the assay.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 20 minutes or Null.
RegenerationShakeRate
Default Calculation: RegenerationShakeRate will be set to 1000 RPM if regeneration steps are included in the assay.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
NeutralizationSolution
The solution that will be used to neutralize the bio-probe surface after regeneration. This solution prevents alteration of the bio-probe surface pH during the regeneration step.
Default Calculation: The NeutralizationSolution will be set to DefaultBuffer if neutralization steps are included in the assay.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
NeutralizationTime
The amount of time for which the bio-probe is immersed in NeutralizationSolution following Regeneration.
Default Calculation: NeutralizationTime will be set to 5 Second if neutralization steps are included in the assay.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 20 minutes or Null.
NeutralizationShakeRate
The plate shake rate while the bio-probe is immersed in NeutralizationSolution following Regeneration.
Default Calculation: NeutralizationShakeRate will be set to 1000 RPM if neutralization steps are included in the assay.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
WashSolution
The solution in which the bio-probe is immersed after neutralization or regeneration, depending on the selection in RegenerationType. This solution prevents interference from the neutralization or regeneration solution.
Default Calculation: The WashSolution will be set to DefaultBuffer if Wash is selected in RegenerationType.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
WashTime
The amount of time for a step in which the bio-probe is immersed in WashSolution following Neutralization.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 20 minutes or Null.
WashShakeRate
The shake rate for a step in which the bio-probe is immersed in WashSolution following Neutralization.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
Probe Loading
LoadingType
Indicates the steps used in the process of loading the immobilized species on the bio-probe. Select all that apply. Load: The bio-probe will be immersed in LoadingSolution prior to sample measurement steps. Activate: The bio-probe will be immersed in ActivationSolution prior to loading. Qunech: The bio-probe will be immersed in QuenchinSolution after loading.
LoadSolution
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
LoadTime
The amount of time for which the bio-probe surface with is functionalized with the immobilized species (in LoadSolution). This allows for modification of the probe surface such that it will be sensitive to the desired analyte. The LoadingTime will limit the step length in the event that a threshold condition is not met.
LoadThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of a Load step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Standard are automatically excluded.
LoadAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the LoadingSolution, and initiates the following assay step. This threshold sets the desired thickness of the loaded bio-layer, and can be used both to ensure sufficient loading and guard against over-saturation of the probe surface. Check Model[Instrument, BioLayerInterferometer] for limits on bio-layer thickness.
LoadThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the LoadingSolution, and initiates the following assay step.
LoadThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to trigger the removal of the bio-probe from the LoadingSolution, and initiate the following assay step.
LoadShakeRate
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
ActivateSolution
Indicates the solution used to enhance the bio-probe surface capacity and affinity for the immobilized species. The bio-probe is immersed in this solution prior to exposure to the LoadingSolution.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
ActivateTime
The amount of time for which the bio-probe is immersed in ActivationSolution prior to loading in LoadingSolution. Activation chemically modifies the probe surface, rendering it more receptive to the immobilized species.
ActivateShakeRate
The speed at which the plate is agitated while the bio-probe is immersed in ActivationSolution prior to loading in LoadingSolution.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
QuenchSolution
The solution used to passivate un-reacted sites after loading the immobilized species by immersion in LoadingSolution. This will reduce the probability of non-selective binding between an analyte and the bio-probe surface.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
QuenchTime
The amount of time for which the bio-probe is immersed in QuenchSolution after being functionalized with the immobilized species. This step passivates un-reacted sites and will reduce the probability of non-selective binding between an analyte and the bio-probe surface.
QuenchShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in QuenchSolution after being functionalized with the immobilized species.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
Kinetics Assay
KineticsReferenceType
Indicates the type of non-sample solutions to be included. Select all that apply. A well containing the selected solution will be measured simultaneously during the association step. If multiple solutions are selected, one well for each solution will be included.
KineticsBaselineBuffer
The solution in which the bio-probes are immersed prior to performing the association step in a kinetics assay. This provides a baseline of the bio-layer thickness prior to analyte association.
Default Calculation: The KineticsBaselineBuffer is set to match the DefaultBuffer if it is informed and the ExperimentType is Kinetics.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
MeasureBaselineTime
The amount of time for which the bio-probe is immersed in KineticsBaselineBuffer directly prior to performing MeasureAssociation.
Default Calculation: MeasureBaselineTime is set to 30 Second if Kinetics is selected as the ExperimentType.
MeasureBaselineShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in KineticsBaselineBuffer directly prior to performing MeasureAssociation.
Default Calculation: MeasureBaselineShakeRate is set to 1000 RPM if Kinetics is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
MeasureAssociationTime
The amount of time for which the bio-probe is immersed in the sample solution to measure analyte association.
Default Calculation: MeasureAssociationTime is set to 15 Minute if Kinetics is selected as the ExperimentType.
MeasureAssociationThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of the Association step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Standard are automatically excluded.
Default Calculation: MeasureAssociationThresholdCriterion is set to All if MeasureAssociationAbsoluteThreshold or MeasureAssociationThresholdSlope is specified.
MeasureAssociationAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the sample solution, and initiate the following assay step. This threshold can be used to prevent excessively long association steps.
MeasureAssociationThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the sample solution, and initiate the following assay step.
MeasureAssociationThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to will trigger the removal of the bio-probe from the sample solution, and initiate the following assay step.
MeasureAssociationShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in the analyte solution.
Default Calculation: MeasureAssociationShakeRate is set to 1000 RPM if Kinetics is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
KineticsDissociationBuffer
Default Calculation: The MeasureDissociationBuffer is set to match the KineticsBaselineBuffer if the ExperimentType is Kinetics.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
MeasureDissociationTime
The amount of time for which the bio-probe is immersed in KineticsDissociationBuffer to measure analyte dissociation.
Default Calculation: MeasureDissociationTime is set to 30 Minute if Kinetics is selected as the ExperimentType.
MeasureDissociationThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of the dissociation step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Standard are automatically excluded.
Default Calculation: MeasureDissociationThresholdCriterion is set to All if MeasureDissociationAbsoluteThreshold or MeasureDissociationThresholdSlope is specified.
MeasureDissociationAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the sample solution, and initiate the following assay step. This threshold can prevent excessively long dissociation steps.
MeasureDissociationThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the sample solution, and initiate the following assay step.
MeasureDissociationThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to will trigger the removal of the bio-probe from the sample solution, and initiate the following assay step.
MeasureDissociationShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in the MeasureDissociationBuffer solution.
Default Calculation: MeasureDissociationShakeRate is set to 1000 RPM if Kinetics is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
Kinetics Sample Preparation
KineticsSampleFixedDilutions
The collection of dilutions that will be performed on each sample to generate dilutions for association measurement.
If the dilutions are prepared in 250 Microliter volumes, they will be performed on the assay plate.
Otherwise, 250 Microliters of each dilution will be transferred to the assay plate.
For Fixed Dilutions, the SampleAmount is the volume of the sample that will be mixed with the DiluentAmount of the Diluent to create a desired concentration.
Default Calculation: This is automatically set Null if Kinetics is not selected in ExperimentType. If Kinetics is selected
it is automatically set to a create a series of 8 solutions which are 250 Microliters each,
with concentrations evenly spaced between the concentration of the sample and 10 fold dilution.
For example, given a 100 Micromolar sample, a series of dilutions with concentrations of
100, 89, 78, 66, 55, 44, 32, 21, 10 Micromolar would be generated.
Programmatic Pattern: (({{GreaterP[0*Microliter], GreaterEqualP[0*Microliter], _String}..} | {{GreaterEqualP[1], _String}..}) | Automatic) | Null
KineticsSampleSerialDilutions
The collection of dilutions that will be performed on each sample to generate dilutions for association measurement.
If the dilutions are prepared in 250 Microliter volumes, they will be performed on the assay plate.
Otherwise, 250 Microliters of each dilution will be transferred to the assay plate.
For Serial Dilutions, the TransferAmount is taken out of the sample and added to a second well with the DiluentAmount of the KineticsSampleDiluent.
It is mixed, then the TransferAmount is taken out of that well to be added to a third well. This is repeated to make samples with the specified SolutionIDs.
Programmatic Pattern: (({GreaterP[0*Microliter], GreaterEqualP[0*Microliter], {_String..}} | {{GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter], _String}..}) | ({GreaterEqualP[2, 1], {_String..}} | {{GreaterEqualP[1, 1], _String}..})) | Null
KineticsSampleDiluent
The solution that is used to dilute the samples to generate the solutions used in KineticsAssociation steps.
Default Calculation: If Kinetics has been selected in ExperimentType, KineticsSampleDiluent will default to DefaultBuffer.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Quantitation Assay
QuantitationParameters
A series of modifications on a basic quantitation experiment which are used inform the assay steps and plate layout. Select all that apply and populate options for the relevant solutions section. StandardCurve: Dilutions of user-specified solution (QuantitationStandard) are used to generate a calibration curve used to quantify analyte concentrations in samples. StandardWell: Simultaneously measure a well containing analyte of known concentration (QuantitationStandard) along with the unknown samples. BlankWell: Simultaneously measure a well of solution not containing the analyte (Blank solution) along with unknown samples. AmplifiedDetection: Performs quantitation measurement in DetectionSolution which amplifies the change in bio-layer thickness, yielding more sensitive detection. This solution is generally an antibody or other substrate that binds to the analyte on the probe surface, thereby increasing the bio-layer thickness CaptureAntibody: Includes a step in which the bio-probe is functionalized with a capture antibody by immersion in CaptureAntibodySolution. SecondaryAntibody: Includes a step in which the bio-probe is treated with another antibody (in SecondaryAntibodySolution) following the first treatment. EnzymeLinked: Exposes the bio-probe to EnzymeSolution for direct measurement, or prior to detection.
Pattern Description: A selection of one or more of StandardCurve, StandardWell, BlankWell, AmplifiedDetection, or EnzymeLinked or Null.
Programmatic Pattern: DuplicateFreeListableP[StandardCurve | StandardWell | BlankWell | AmplifiedDetection | EnzymeLinked] | Null
QuantitationStandard
Indicates a solution which is used to generate a standard curve for quantitation of the analyte concentration in the input samples.
Default Calculation: The QuantitionStandard will be set to the same value as the Standard if Quantitation is selected in ExperimentType and if the Standard has been user specified.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
QuantitationStandardWell
Default Calculation: The QuantitionStandardWell will be set to the same value as the QuantitationStandard if Quantitation is selected in ExperimentType and if the QuantitationStandard has been user specified.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
QuantitateTime
The amount of time for which the bio-probe is immersed in the sample solution to perform a quantitation measurement. This time will also apply to steps measuring the QuantitationStandard solutions, if requested in QuantititationParameters.
Default Calculation: QuantitateSampleMeasurementTime is set to 5 minutes if Quantitation is selected as the ExperimentType.
QuantitateShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in the sample solution to perform a quantitation measurement. This will also apply to steps measuring the QuantitationStandard solutions, if requested in QuantititationParameters.
Default Calculation: QuantitateSampleMeasurementShakeRate is set to 1000 RPM if Quantitation is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
AmplifiedDetectionSolution
A solution containing a species which binds to the immobilized analyte on the bio-probe surface, thereby increasing the thickness of the bio-layer. This solution can improve the detection of an analyte in quantitation experiments, and is most commonly used when detecting enzymes.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
AmplifiedDetectionTime
The amount of time for an amplified quantitation measurement step, in which the bio-probe is immersed in the AmplifiedDetectionSolution.
Default Calculation: AmplifiedDetectionTime is set to 5 minutes if AmplifiedDetection is selected in QuantitationParameters.
AmplifiedDetectionShakeRate
The speed at which the assay plate is shaken for an amplified quantitation measurement step, in which the bio-probe is immersed in the DetectionSolution.
Default Calculation: AmplifiedDetectionShakeRate is set to 1000 RPM if AmplifiedDetection is selected in QuantitationParameters.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
QuantitationEnzymeSolution
Indicates a solution containing enzyme used to amplify quantitation results. The probe is immersed in QuantitationEnzymeSolution after immersion in the analyte solution.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
QuantitationEnzymeBuffer
Default Calculation: QuantitationEnzymeBuffer will be set to DefaultBuffer if QuantitaionEnzyme is selected in QuantitationParameters.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
QuantitationEnzymeTime
The amount of time which the bio-probe is immersed in an enzyme solution (QuantitationEnzymeSolution). This step is performed after bio-probe immersion in the sample solutions.
Default Calculation: QuantitationEnzymeTime is set to 5 minutes if QuantitaionEnzyme is selected in QuantitationParameters.
QuantitationEnzymeShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in an enzyme solution (QuantitationEnzymeSolution). This step is performed after bio-probe immersion in the sample solutions.
Default Calculation: QuantitationEnzymeShakeRate is set to 1000 RPM if QuantitaionEnzyme is selected in QuantitationParameters.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
Quantitation Standard Preparation
QuantitationStandardFixedDilutions
The collection of dilutions that will be performed on the QuantitationStandard to generate a standard curve for a Quantitation experiment.
If the dilutions are prepared in 250 Microliter volumes, they will be performed on the assay plate.
Otherwise, 250 Microliters of each dilution will be transferred to the assay plate.
For Fixed Dilution Volumes, the SampleAmount is the volume of the sample that will be mixed with the DiluentAmount of the Diluent to create a desired concentration.
For Fixed Dilution Factors, an appropriate amount of sample will mixed with diluent to achieve a solution with the requested volume and concentration.
The SolutionIDs are used to indicate a unique temporary name for each dilution that is referred to in the protocol object.
Default Calculation: This is automatically set to Null if QuantitationStandard is set to Null. In all
other cases it is automatically set to a create a series of dilutions of the QuantitationStandard using dilution factors of 2, 4, 8, 16, 32, 64, and 128 and diluting with QuantiationDiluent.
Programmatic Pattern: (({{GreaterP[0*Microliter], GreaterP[0*Microliter], _String}..} | {{GreaterEqualP[1], _String}..}) | Automatic) | Null
QuantitationStandardSerialDilutions
The collection of dilutions that will be performed on the QuantitationStandard to generate a standard curve for a Quantitation experiment.
For volume based Serial Dilutions, the TransferAmount is taken out of the QuantitationStandard and added to a second well with the DiluentAmount of the QuantitationStandardDiluent.
It is mixed, then the TransferAmount is taken out of that well to be added to a third well. This is repeated to the solutions labeled by SolutionIDs.
For dilution factor based Serial Dilutions, the appropriate amount of solution will be added to a diluent to achieve the solutions with desired dilution factor and final volumes of > 250 uL. The SolutionIDs are used to indicate a unique temporary name for each dilution that is referred to in the protocol object.
Programmatic Pattern: (({GreaterP[0*Microliter], GreaterEqualP[0*Microliter], {_String..}} | {{GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter], _String}..}) | ({GreaterEqualP[2, 1], {_String..}} | {{GreaterEqualP[1, 1], _String}..})) | Null
QuantitationStandardDiluent
The solution that is used to dilute the QuantitationStandard to generate the solutions used in the quantitation standard curve.
Default Calculation: If the a dilution series has been specified, QuantitationStandardDiluent will default to DefaultBuffer.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Epitope Binning Assay
BinningType
Indicates the assay configuration for EpitopeBinning. Sandwich: The first antibody is bound to the bio-probe surface. The surface is first exposed to the target antigen (BinningAntigen), then to solutions of competing antibodies. Tandem: The target antigen species is immobilized on the bio-probe surface. The surface is then exposed to a pair of competing antibodies sequentially. Premix: An antibody is bound to the bio-probe surface. The bio-probe is then exposed to a premixed solution of antigen and competing antibody.
Default Calculation: If EpitopeBinning is selected in ExperimentType, BinningType will default to Sandwich, otherwise it will default to Null.
BinningControlWell
During the Antibody loading step, one well of Blank solution will be included. This will reduce the maximum number of antibodies from 8 to 7.
Default Calculation: BinningControlWell defaults to True if EpitopeBinning is selected in ExperimentType, otherwise it will default to Null.
LoadAntibodyTime
For Sandwich-type assays: The amount of time for which the bio-probe surface with is saturated with bound antibody. For Tandem-type assays: The amount of time for which the antigen coated bio-probe surface with is immersed in the first antibody solution.
Default Calculation: LoadAntibodyTime is set to 10 minutes if EpitopeBinning is selected as the ExperimentType.
LoadAntibodyThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of an assay step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Standard are automatically excluded.
Default Calculation: LoadAntibodyThresholdCriterion is set to All if LoadAntibodyAbsoluteThreshold or LoadAntibodyThresholdSlope is specified.
LoadAntibodyAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the antibody solution, and initiate the following assay step. This threshold can be used to ensure an appropriate amount of antibody association, and prevent over-saturation of the probe surface.
LoadAntibodyThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the antibody solution, and initiate the following assay step.
LoadAntibodyThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to will trigger the removal of the bio-probe from the antibody solution, and initiate the following assay step.
LoadAntibodyShakeRate
For Sandwich-type assays: The speed at which the assay plate is shaken while the bio-probe surface with is saturated with bound antibody. For Tandem-type assays: The speed at which the assay plate is shaken while the antigen coated bio-probe surface with is immersed in the first antibody solution.
Default Calculation: LoadAntibodyShakeRate is set to 1000 RPM if EpitopeBinning is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
BinningQuenchSolution
Indicates a solution used to quench unreacted sites on the probe surface after antibody loading. Immersion of the bio-probe in this solution can help prevent non-selective binding in subsequent assay steps.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
BinningQuenchTime
The amount of time for which the bio-probe is immersed in BinningQuenchSolution, which blocks unreacted sites. This option will override any assignment made in Loading Information.
Default Calculation: BinningQuenchTime is set to 5 minutes if EpitopeBinning is selected as the ExperimentType.
BinningQuenchShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in BinningQuenchSolution, which blocks unreacted sites. This option will override any assignment made in Loading Information.
Default Calculation: BinningQuenchShakeRate is set to 1000 RPM if EpitopeBinning is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
BinningAntigen
Indicates a solution used in the Tandem and Sandwich type assays to populate the bio-probe surface with the target antigen.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
LoadAntigenTime
For Sandwich-type assays: The amount of time while the antibody functionalized bio-probe surface is exposed to BinningAntigen. For Tandem-type assays: The amount of time while the bio-probe surface is saturated with antigen in BinningAntigen solution.
Default Calculation: LoadAntigenTime is set to 10 minutes if EpitopeBinning is selected as the ExperimentType.
LoadAntigenThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of an antigen association step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Standard are automatically excluded.
Default Calculation: LoadAntigenThresholdCriterion is set to All if LoadAntigenAbsoluteThreshold or LoadAntigenThresholdSlope is specified.
LoadAntigenAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the antigen solution, and initiate the next assay step. This threshold can ensure an appropriate amount of antigen association, preventing oversaturation of the probe surface.
LoadAntigenThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the antigen solution, and initiate the next assay step.
LoadAntigenThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to will trigger the removal of the bio-probe from the antibody solution, and initiate the next assay step.
LoadAntigenShakeRate
For Sandwich-type assays: The speed at which the assay plate is shaken for a step in which the antibody functionalized bio-probe surface is exposed to BinningAntigen. For Tandem-type assays: The speed at which the assay plate is shaken for a step in which the bio-probe surface is saturated with antigen in BinningAntigen.
Default Calculation: LoadAntigenShakeRate is set to 1000 RPM if EpitopeBinning is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
CompetitionBaselineBuffer
The solution in which the bio-probe is immersed prior to performing the competition step of an EpitopeBinning assay.
Default Calculation: The CompetitionBaselineBuffer is set to match the DefaultBuffer if the ExperimentType is EpitopeBinning.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
CompetitionBaselineTime
The amount of time while the bio-probe is immersed in BinningBaselineBuffer solution prior to performing the competition step of a EpitopeBinning assay.
Default Calculation: CompetitionBaselineTime is set to 30 seconds if EpitopeBinning is selected as the ExperimentType.
CompetitionBaselineShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in CompetitionBaselineBuffer solution prior to performing the competition step of a EpitopeBinning assay.
Default Calculation: CompetitionBaselineShakeRate is set to 1000 RPM if EpitopeBinning is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
CompetitionTime
The amount of time while the bio-probe is immersed in a competing antibody (sample) or premixed antibody/antigen solution to observe competitive binding.
Default Calculation: CompetitionTime is set to 10 minutes if EpitopeBinning is selected as the ExperimentType.
CompetitionShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in a competing antibody (sample) or premixed antibody/antigen solution to observe competitive binding.
Default Calculation: CompetitionShakeRate is set to 1000 RPM if EpitopeBinning is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
CompetitionThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of an assay step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Standard are automatically excluded.
Default Calculation: CompetitionThresholdCriterion is set to Single if CompetitionAbsoluteThreshold or CompetitionThresholdSlope is specified.
CompetitionAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the antibody solution, and initiate the following assay step. This threshold can be used prevent excessively long step times.
CompetitionThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the sample solution, and initiates the following assay step. This can be used to ensure that the probe surface reaches an equilibrium condition.
CompetitionThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to will trigger the removal of the bio-probe from the sample solution, and initiates the following assay step.
PreMixSolutions
The mixture of antibody solutions (the sample solutions) and antigen solution (BinningAntigen) which compose the PreMixSolutions.
The solutions can be combined with PreMixDiluent to adjust the final concentration of the PreMixSolutions.
Default Calculation: This is automatically set Null if PreMix is not selected in BinningExperimentType. If PreMix is selected
it is automatically set to mix 100 Microliters of the sample solution with 100 Microliters of the BinningAntigen.
Pattern Description: {SampleAmount, BinningAntigenAmount, PreMixDiluentAmount, SolutionIDs} or Null.
Programmatic Pattern: ({GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter], _String} | Automatic) | Null
PreMixDiluent
Default Calculation: If PreMix has been selected in BinningType, PreMixDiluent will be set to DefaultBuffer.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Assay Development
DevelopmentType
Indicates which step or solution is being optimized or investigated. All assays will include an association step (in the input sample solution) and a dissociation step (in DefaultBuffer or TestBufferSolutions). ScreenLoading: Find the best loading condition or immobilized species from a list of TestLoadingSolutions. ScreenInteraction: Test interaction of pairs of immobilized and solution species using the TestInteractionSolutions. These solutions will be used in the load step, and are index matched to the sample input. ScreenBuffer: Perform measurements using a list of TestBufferSolutions. ScreenRegeneration: Perform regeneration using a series of TestRegenerationSolutions. ScreenActivation: Test activation conditions prior to a LoadSurface step, using different activation TestActivationSolutions. ScreenDetectionLimit: Perform identical measurements on increasingly dilute solutions to determine the detection limit for a given analyte.
Default Calculation: If AssayDevelopment is selected in ExperimentType, the DevelopmentType is set to ScreenDetectionLimit.
Pattern Description: ScreenLoading, ScreenInteraction, ScreenBuffer, ScreenRegeneration, ScreenActivation, or ScreenDetectionLimit or Null.
Programmatic Pattern: ((ScreenLoading | ScreenInteraction | ScreenBuffer | ScreenRegeneration | ScreenActivation | ScreenDetectionLimit) | Automatic) | Null
DevelopmentReferenceWell
Indicates if a well containing Blank, Standard or both should be measured in parallel with the samples in the developmentAssociation step.
DevelopmentBaselineTime
The amount of time a bio-probe is immersed in buffer (DefaultBuffer or TestBufferSolutions) solution to record a baseline.
Default Calculation: DevelopmentBaselineTime is set to 30 seconds if AssayDevelopment is selected as the ExperimentType.
DevelopmentBaselineShakeRate
The speed at which the assay plate is shaken while a bio-probe is immersed in buffer (DefaultBuffer or TestBufferSolutions) solution to record a baseline.
Default Calculation: DevelopmentBaselineShakeRate is set to 1000 RPM if AssayDevelopment is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
DevelopmentAssociationTime
The amount of time which the bio-probe is immersed in sample solution to measure analyte association.
Default Calculation: AssayDevelopmentAssociationTime is set to 5 minutes if AssayDevelopment is selected as the ExperimentType.
DevelopmentAssociationThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of an association step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Standard are automatically excluded.
Default Calculation: DevelopmentAssociationThresholdCriterion is set to All if DevelopmentAssociationAbsoluteThreshold or DevelopmentAssociationThresholdSlope is specified.
DevelopmentAssociationAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the sample solution, and initiate the next assay step. This threshold can prevent excessively long association steps.
DevelopmentAssociationThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the sample solution, and initiate the next assay step.
DevelopmentAssociationThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to will trigger the removal of the bio-probe from the sample solution, and initiate the next assay step.
DevelopmentAssociationShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in sample solution to measure analyte association.
Default Calculation: AssayDevelopmentAssociationShakeRate is set to 1000 RPM if AssayDevelopment is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
DevelopmentDissociationTime
The amount of time for which the bio-probe is immersed in DefaultBuffer or TestBufferSolutions during analyte dissociation.
Default Calculation: AssayDevelopmentDissociationTime is set to 30 minutes if AssayDevelopment is selected as the ExperimentType.
DevelopmentDissociationThresholdCriterion
Indicates if the threshold condition for change in bio-layer thickness which will trigger the completion of an dissociation step must be met by any single well, or all of the wells measured in the step. Wells containing a secondary solution such as a Blank or Control are automatically excluded.
Default Calculation: DevelopmentDissociationThresholdCriterion is set to All if DevelopmentDissociationAbsoluteThreshold or DevelopmentDissociationThresholdSlope is specified.
DevelopmentDissociationAbsoluteThreshold
The change in bio-layer thickness that will trigger the removal of the bio-probe from the buffer solution, and initiate the next assay step. This threshold can be used to prevent excessively long dissociation steps.
DevelopmentDissociationThresholdSlope
The rate of change in bio-layer thickness that will trigger the removal of the bio-probe from the buffer solution, and initiate the next assay step.
DevelopmentDissociationThresholdSlopeDuration
The amount of time that a given rate of change in bio-layer thickness must be exceeded to will trigger the removal of the bio-probe from the buffer solution, and initiate the next assay step.
DevelopmentDissociationShakeRate
The speed at which the assay plate is shaken while the bio-probe is immersed in DefaultBuffer or TestBufferSolutions during analyte dissociation.
Default Calculation: AssayDevelopmentDissociationShakeRate is set to 1000 RPM if AssayDevelopment is selected as the ExperimentType.
Pattern Description: Greater than or equal to 100 revolutions per minute and less than or equal to 1500 revolutions per minute or Null.
Assay Development Solutions
DetectionLimitSerialDilutions
The collection of dilutions that will be performed on each sample to generate dilutions to determine the limit of detection for one or more analytes.
If the dilutions are prepared in 250 Microliter volumes, they will be performed on the assay plate.
Otherwise, 250 Microliters of each dilution will be transferred to the assay plate.
For Serial Dilutions, the TransferAmount is taken out of the sample and added to a second well with the DiluentAmount of the DetectionLimitDiluent.
It is mixed, then the TransferAmount is taken out of that well to be added to a third well. This is repeated to make each of the solutions labeled by SolutionIDs.
Default Calculation: This is automatically set Null if DetectionLimit is not selected in DevelopmentType. If DetectionLimit is selected
it is automatically set to a create a series of 6 solutions which are 250 Microliters each,
using serial dilution of 50 Microliters of sample into 250 Microliters of DetectionLimitDiluent.
Programmatic Pattern: ((({GreaterP[0*Microliter], GreaterEqualP[0*Microliter], {_String..}} | {{GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter], _String}..}) | ({GreaterEqualP[2, 1], {_String..}} | {{GreaterEqualP[1, 1], _String}..})) | Automatic) | Null
DetectionLimitFixedDilutions
The collection of dilutions that will be performed on each sample to generate dilutions to determine the limit of detection for one or more analytes.
If the dilutions are prepared in 250 Microliter volumes, they will be performed on the assay plate.
Otherwise, 250 Microliters of each dilution will be transferred to the assay plate.
For Fixed Dilution Volumes, the SampleAmount is the volume of the sample that will be mixed with the DiluentAmount of the DetectionLimitDiluent to create a desired concentration.
For Fixed Dilution Factors, the appropriate amount of sample volume is mixed with diluent to create a solution with the desired dilution factor and volume.
Programmatic Pattern: ({{GreaterP[0*Microliter], GreaterP[0*Microliter], _String}..} | {{GreaterEqualP[1], _String}..}) | Null
DetectionLimitDiluent
The solution that is used to dilute the samples to generate a set of sample which can be used to establish a limit of detection for a given bio-probe/analyte pairing.
Default Calculation: If DetectionLimit has been selected in DevelopmentType, DetectionLimitDiluent will default to DefaultBuffer.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
TestInteractionSolutions
The list of solutions which are used to load the bio-probe surface for pair-wise interaction screening. This list is required if ScreenInteraction is selected.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
TestBufferSolutions
Pattern Description: List of one or more an object of type or subtype Object[Sample] or Model[Sample] or a prepared sample entries or Null.
TestRegenerationSolutions
The list of solutions to be used to return the probe to a measurement-ready condition. This list is required if ScreenRegeneration is selected.
Pattern Description: List of one or more an object of type or subtype Object[Sample] or Model[Sample] or a prepared sample entries or Null.
TestLoadingSolutions
The list of solutions in which the bio-probe will be immersed to load the immobilized species. This list is required if ScreenLoading is selected.
Pattern Description: List of one or more an object of type or subtype Object[Sample] or Model[Sample] or a prepared sample entries or Null.
TestActivationSolutions
The list of solutions in which the bio-probe will be immersed in prior to loading the immobilized species. This list is required if ScreenActivation is selected.
Pattern Description: List of one or more an object of type or subtype Object[Sample] or Model[Sample] or a prepared sample entries or Null.
Assay Primitives
AssaySequencePrimitives
The sequence of assay steps which will apply to each input sample. This sequence will be repeated the necessary number of times to perform the requested experiments, which the exception of Activation/Loading/Quench sequences, which will not be repeated if Regenerate steps are included. Analytes/Solution/Buffers: The solution(s) required for the assay step. Controls: Solutions (usually a blank or control) which will occupy one well in the assay plate, and are measured in parallel with the primary solution. Time: The amount of time for which the probe is immersed in the solution, if a threshold condition is not met, this time will limit the step length. Threshold: Select between Absolute (based on the total change in bio-layer thickness) and Slope (based on the rate of change in the bio-layer thickness). AbsoluteThreshold: The change in bio-layer thickness which will trigger the next assay step to begin. This can be used to ensure an appropriate amount of association to the probe surface. ThresholdType: Indicates if the threshold conditions must be met by any single well or all of the wells containing the primary solution. ThresholdSlope: The change in bio-layer thickness over time. ThresholdSlopeDuration: The amount of time for which the change in bio layer thickness over time must be less than that the ThresholdSlope to trigger the next step. ShakeRate: The rate at which the assay plate is shaken during this step.
Default Calculation: The order, type, and contents of the primitives are determined from the options. For example, setting options ExperimentType -> Kinetics, and LoadingType -> Loading would generate AssaySequencePrimitives of {Equilibrate,Load,Baseline,MeasureAssociation,MeasureDissociation}, with the solution, step times, thresholds, and shake-rate as specified in the options.
Pattern Description: List of one or more a primitive with head MeasureBaseline, Equilibrate, ActivateSurface, LoadSurface, Quantitate, Quench, Regenerate, Neutralize, Wash, MeasureAssociation, or MeasureDissociation entries or Null.
ExpandedAssaySequencePrimitives
The exact sequence of assay steps which will be performed for this bio-layer interferometry experiment, grouped by steps that will be performed with the same probe.
Default Calculation: The ExpandedAssaySequencePrimitives are based on the AssaySequencePrimitives as defined directly by the user or by user input options.
Pattern Description: List of one or more list of one or more a primitive with head MeasureBaseline, Equilibrate, ActivateSurface, LoadSurface, Quantitate, Quench, Regenerate, Neutralize, Wash, MeasureAssociation, or MeasureDissociation entries entries.
RepeatedSequence
The sequence of steps that are repeated by each bio-probe when regeneration is requested. For example, if the AssaySequencePrimitives -> {Load, Quantitate, Regenerate}, RepeatedSequence -> {Load, Quantitate, Regenerate} would return the probe to its original state, while RepeatedSequence -> {Quantitate, Regenerate} would return the surface as functionalized in the Load step.
Default Calculation: RepeatedSequence resolves to the subset of AssaySequencePrimitives names which occur for each sample following the initial measurement. RepeatedSequence resolves to Null unless there is a Regenerate step in AssaySequencePrimitives
Pattern Description: List of one or more {Equilibrate, Wash, ActivateSurface, Quench, LoadSurface, Regenerate, Neutralize, MeasureBaseline, MeasureAssociation, MeasureDissociation, Quantitate} entries or Null.
Model Input
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Programmatic Pattern: ((Null | (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All)) | Automatic) | Null
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSamplePreparation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentratedBufferDilutionFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Kinetics
Quantitation
Quantify the amount of an analyte in a given sample using direct measurement and a calibration curve:
Quantify the amount of an analyte in a given sample using indirect (amplified) detection. This is useful in cases where analyte association does not appreciably change the biolayer thickness:
Quantify the amount of an analyte in a given sample by specifying a standard curve with fixed dilutions in the form of {Standard Volume, Diluent Volume, Temporary Solution Name}:
Quantify the amount of an analyte in a given sample by specifying a standard curve with serial dilutions in the form of {Dilution Factor, Number Of Dilutions, {Temporary Solution Names}}:
Epitope Binning
Screening
Warnings and Errors
Messages (35)
BLIBinningAntigenStorageConditionMismatch (1)
BLIConflictingDevelopmentDilutions (1)
BLIConflictingKineticsDilutions (1)
BLIDuplicateDilutionNames (2)
BLIForbiddenRepeatedSequence (1)
BLILoadSolutionStorageConditionMismatch (1)
BLIMissingDevelopmentDiluents (1)
BLIMissingKineticsDiluent (1)
BLIMissingPreMixDiluent (1)
BLIProbeApplicationMismatch (1)
BLIQuantitationEnzymeStorageConditionMismatch (1)
BLIQuantitationStandardStorageConditionMismatch (1)
BLIRepeatedSequenceMismatch (1)
BLIStandardStorageConditionMismatch (1)
BLITestInteractionSolutionsStorageConditionMismatch (1)
BLITestLoadingSolutionsStorageConditionLengthMismatch (1)
BLITestLoadingSolutionsStorageConditionMismatch (1)
BLIUnspecifiedQuantitationStandard (1)
ObjectDoesNotExist (6)
Do NOT throw a message if we have a simulated container but a simulation is specified that indicates that it is simulated:
Do NOT throw a message if we have a simulated sample but a simulation is specified that indicates that it is simulated:
Throw a message if we have a container that does not exist (ID form):
Throw a message if we have a container that does not exist (name form):
Throw a message if we have a sample that does not exist (ID form):
Throw a message if we have a sample that does not exist (name form):
UnusedBLIOptionValuesAssayDevelopment (1)
UnusedBLIOptionValuesEpitopeBinning (1)
UnusedBLIOptionValuesKinetics (1)
UnusedBLIOptionValuesQuantitation (1)
UnusedOptionValuesBLIPrimitiveInput (2)
Possible Issues
Sample evaporation
Assays longer than 4 hours may exhibit excessive evaporation which can impact experimental results. In this case, it is recommended that a plate cover be used to limit evaporation.
Insufficient well volume
Well volumes below 200 uL may result in sensor failure or inaccurate data due to incomplete probe immersion. Evaporation can be prevented by using the AssayPlateCover option.
Insufficient probe hydration
Probes should be immersed in buffer solution for at least 10 minutes prior to the assay beginning. Insufficient probe equilibration can lead to sensor failure. The use of the ProbeRackEquilibration option is highly recommended and will generally prevent this issue.
Baseline drift
Nonspecific binding
Nonspecific binding may occur in cases where the surface is not properly passivated or a suboptimal buffer is selected. The suggested remedy is to screen buffers or quench solutions to find a suitable solution.
Overloaded probe surface
Last modified on Wed 8 Oct 2025 12:51:05