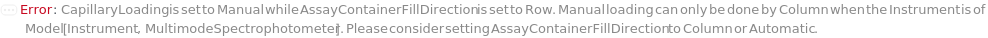General
AssayType
The Dynamic Light Scattering (DLS) assay that is run. SizingPolydispersity makes a single DLS measurement that provides information about the size and polydispersity (defined as the ratio of mass-average molar mass to number-average molar mass) of particles in the input samples. ColloidalStability makes DLS measurements at various dilutions of a sample below 25 mg/mL to calculate the diffusion interaction parameter (kD) and the second virial coefficient (B22 or A2), and does the same for a sample of mass concentration 20-100 mg/mL to calculate the Kirkwood-Buff Integral (G22) at each dilution concentration; Static Light Scattering (SLS) measurements can be used to calculate A2 and the molecular weight of the analyte. MeltingCurve makes DLS measurements over a range of temperatures in order to calculate melting temperature (Tm), temperature of aggregation (Tagg), and temperature of onset of unfolding (Tonset). IsothermalStability makes multiple DLS measurements at a single temperature over time in order to probe stability of the analyte at a particular temperature.
Default Calculation: The AssayType is set to IsothermalStability if any of the options in the "Isothermal Stability" category are specified, to MeltingCurve if any of the options in the "Melting Curve" category are defined, to ColloidalStability if Dilution-related options are specified, and to SizingPolydispersity otherwise.
Pattern Description: SizingPolydispersity, IsothermalStability, MeltingCurve, or ColloidalStability.
Programmatic Pattern: DynamicLightScatteringAssayTypeP | Automatic
AssayFormFactor
Indicates if the sample is loaded in capillary strips, which are utilized by a Multimode Spectrophotometer, or a standard plate which is utilized by a DLS Plate Reader.
Default Calculation: AssayFormFactor is set to Capillary if the Instrument model is set to Model[Instrument,MultimodeSpectrophotometer,"Uncle"] and Plate if Instrument is set to Model[Instrument,DLSPlateReader,"DynaPro"]. If neither AssayFormFactor nor Instrument is defined, AssayFormFactor is set to Capillary if SampleVolume is set to 25 uL and Object[Instrument,DLSPlateReader,"DynaPro" if SampleVolume is set to greater than 25 uL.
Pattern Description: Capillary or Plate.
Programmatic Pattern: (Capillary | Plate) | Automatic
Instrument
The instrument used for this experiment. Options comprise Model[Instrument,MultimodeSpectrophotometer,"Uncle"], or any instruments of that model, which uses a capillary form factor and can perform fluorescence as well as light scattering experiments assays, and Model[Instrument,DLSPlateReader,"DynaPro"], or any instruments of that model, which uses a plate form factor and performs only light scattering experiments.
Default Calculation: Instrument is set to Model[Instrument,MultimodeSpectrophotometer,"Uncle"] if AssayFormFactor is set to Capillary and Model[Instrument,DLSPlateReader,"DynaPro"] if AssayFormFactor is set to Plate. If neither AssayFormFactor nor Instrument is defined, Instrument is set to Model[Instrument,MultimodeSpectrophotometer,"Uncle"] if SampleVolume is set to 25 uL or less and Model[Instrument,DLSPlateReader,"DynaPro"] if SampleVolume is set to greater than 25 uL.
Pattern Description: An object of type or subtype Object[Instrument, MultimodeSpectrophotometer], Model[Instrument, MultimodeSpectrophotometer], Object[Instrument, DLSPlateReader], or Model[Instrument, DLSPlateReader]
Programmatic Pattern: ObjectP[{Object[Instrument, MultimodeSpectrophotometer], Model[Instrument, MultimodeSpectrophotometer], Object[Instrument, DLSPlateReader], Model[Instrument, DLSPlateReader]}] | Automatic
AssayContainers
The capillary strips or plates that the samples in this experiment are assayed in. Options comprise a list of one of Model[Container, Plate, "96 well Flat Bottom DLS Plate"], Model[Container, Plate, "96 well Clear Flat Bottom DLS Plate"], Model[Container, Plate, "384-well Aurora Flat Bottom DLS Plate"], each of which use a plate form factor, and a list of one, two, or three of Model[Container, Plate, CapillaryStrip, "Uncle 16-capillary strip"], which uses a capillary form factor, or any objects of those models. If AssayFormFactor is Capillary, it is strongly recommended to allow AssayContainers to resolve automatically.
Default Calculation: When AssayFormFactor is set to Plate, AssayContainers is set to {Model[Container, Plate, "96 well Flat Bottom DLS Plate"]} if SampleVolume is set to more than 30 Microliter and there are fewer than 96 wells required for the assay, and to {Model[Container, Plate, "384-well Aurora Flat Bottom DLS Plate"]} otherwise. When AssayFormFactor is set to Capillary, AssayContainers is set to one or more instances of Model[Container, Plate, CapillaryStrip, "Uncle 16-capillary strip"], depending on the number of wells required for the assay.
Pattern Description: List of one or more an object of type or subtype Object[Container, Plate] or Model[Container, Plate] or a prepared sample entries.
Programmatic Pattern: {(ObjectP[{Object[Container, Plate], Model[Container, Plate]}] | _String)..} | Automatic
Sample Loading
SampleLoadingPlate
The container into which input samples are transferred (or in which input sample dilutions are performed when AssayType is ColloidalStability) before centrifugation and transfer into the AssayContainer(s) for DLS measurement.
Default Calculation: The SampleLoadingPlate is set to Null when AssayFormFactor is Plate and to Model[Container, Plate, "96-well PCR Plate"] when AssayFormFactor is Capillary.
Pattern Description: An object of type or subtype Object[Container, Plate] or Model[Container, Plate] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Container, Plate], Model[Container, Plate]}] | _String) | Automatic) | Null
NumberOfReplicates
The number of times the specified Dynamic Light Scattering (DLS) assay is run on each input sample. In practice, this refers to the number of wells of the AssayContainer(s) that each input sample occupies. For example, ExperimentDynamicLightScattering[{input1, input2}, NumberOfReplicates->2] is equivalent to ExperimentDynamicLightScattering[{input1, input1, input2, input2}]. When AssayType is SizingPolydispersity, IsothermalStability, and MeltingCurve, NumberOfReplicates refers to the number of wells for each distinct input sample. When AssayType is ColloidalStability, NumberOfReplicates refers to the replicates of each dilution concentration of each sample. At least two replicates of each dilution concentration are required for ColloidalStability, and three are recommended.
Default Calculation: Automatically set to 3 if AssayType is ColloidalStability and 2 otherwise.
Pattern Description: Greater than or equal to 1 and less than or equal to 384 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 384, 1] | Automatic) | Null
AssayContainerFillDirection
WhenAssayFormFactor is Capillary, controls how the capillary strip AssayContainers are filled. Column indicates that all 16 wells of an AssayContainer capillary strip (Model[Container, Plate, CapillaryStrip,"Uncle 16-capillary strip"]) are filled with input samples or sample dilutions before starting to fill a second capillary strip (up to 3 strips, 48 wells). Row indicates that one well of each capillary strip is filled with input samples or sample dilutions before filling the second well of each strip. Setting the AssayContainerFillDirection to Column will minimize the number of capillary strips needed for the experiment, while Row will always use three capillary strips if there are more than two input samples. Setting this option to Row will ensure that replicate dilution concentration measurements in ColloidalStability occur in separate capillary strips. When AssayFormFactor is Plate, controls the direction the AssayContainer is filled: either Row, Column, SerpentineRow, or SerpentineColumn.
Pattern Description: Row, Column, SerpentineRow, or SerpentineColumn or Null.
Programmatic Pattern: (Row | Column | SerpentineRow | SerpentineColumn) | Null
SampleVolume
When AssayType is SizingPolydispersity, MeltingCurve, or IsothermalStability, the SampleVolume is the amount of each input sample that is transferred into the SamplePreparationPlate before the SamplePreparationPlate is centrifuged and 10 uL of each sample is transferred into the AssayContainer(s) for DLS measurement. When the AssayType is ColloidalStability, the amount of input sample required for the experiment is specified with either the DilutionCurve or SerialDilutionCurve option.
Default Calculation: If the AssayType is SizingPolydispersity, MeltingCurve, or IsothermalStability, when the AssayFormFactor is Capillary, SampleVolume is set to 15 uL; when the AssayFormFactor is Plate, SampleVolume is set to 30 uL (for a 384-well plate) or 100 uL (for a 96-well plate); and set to Null otherwise.
Pattern Description: Greater than or equal to 15 microliters and less than or equal to 100 microliters or Null.
Programmatic Pattern: (RangeP[15*Microliter, 100*Microliter] | Automatic) | Null
Temperature
The temperature to which the incubation chamber is set prior to detection.
Default Value: 25 degrees Celsius
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 95 degrees Celsius.
Programmatic Pattern: RangeP[4*Celsius, 95*Celsius]
EquilibrationTime
The length of time for which the samples are held in the chamber which is incubated at the Temperature before the first DLS measurement is made, in order to warm or cool the samples to Temperature.
Pattern Description: Greater than or equal to 1 second and less than or equal to 24 hours.
Programmatic Pattern: RangeP[1*Second, 24*Hour]
CapillaryLoading
The loading method for capillaries. When set to Robotic, capillaries are loaded by liquid handler. When set to Manual, capillaries are loaded by a multichannel pipette. Each method presents specific mechanical challenges due to the difficulty of precise loading.
Default Calculation: When AssayFormFactor is Capillary, set to Manual. When AssayFormFactor is Plate, set to Null.
Pattern Description: Robotic or Manual or Null.
Programmatic Pattern: ((Robotic | Manual) | Automatic) | Null
WellCover
When AssayFormFactor is Plate, determines what covering will be used for wells. Available coverings are plate seal, and oil; other covers (e.g. lids) have not yet been evaluated for their effects on light scattering optics.
Default Calculation: When AssayFormFactor is Plate, automatically set to Model[Sample,"Silicone Oil"]. When AssayFormFactor is Capillary, automatically set to Null.
Pattern Description: An object of type or subtype Object[Item, PlateSeal], Model[Item, PlateSeal], Object[Sample], or Model[Sample] or a prepared sample or None or Null.
Programmatic Pattern: (((ObjectP[{Object[Item, PlateSeal], Model[Item, PlateSeal], Object[Sample], Model[Sample]}] | _String) | None) | Automatic) | Null
WellCoverHeating
When WellCover is Model[Item,PlateSeal] or any object of that model, indicates if the plate seal is heated.
Default Calculation: When AssayFormFactor is Plate and WellCover is set to a Model[Item,PlateSeal] or Object[Item,PlateSeal], automatically set to True. When AssayFormFactor is Capillary or WellCover is set to anything other than Model[Item,PlateSeal] or Object[Item,PlateSeal], automatically set to Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Sample Dilution
ReplicateDilutionCurve
Indicates if a NumberOfReplicates number of StandardDilutionCurves or SerialDilutionCurves are made for each input sample. When ReplicateDilutionCurve is True, the replicate DLS measurements for ColloidalStability assay are made from the same concentration of each of the StandardDilutionCurves or SerialDilutionCurves created from a given input sample. When ReplicateDilutionCurve is False, the replicate DLS measurements for the ColloidalStability assay are made from aliquots of a given concentration of the DilutionCurve or SerialDilutionCurve.
Default Calculation: If the AssayType is ColloidalStability, ReplicateDilutionCurve is set to True if the AssayFormFactor is Plate, and to False if AssayFormFactor is Capillary. If the AssayType is not ColloidalStability, ReplicateDilutionCurve is set to Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
StandardDilutionCurve
The collection of dilutions that are performed on each input sample in the SampleLoadingPlate or AssayContainer. For Fixed Dilution Volume Dilution Curves, the Sample Amount is the volume of the sample that is mixed with the Buffer to the Total Volume to create a desired concentration. For Fixed Target Concentration Dilution Curves, the Target Concentration is the desired final concentration of analyte after dilution of the input samples with the Buffer.
Default Calculation: The Dilution is set to Null if the AssayType is SizingPolydispersity, IsothermalStability, or MeltingCurve, or if the SerialDilutionCurve option is specified. Otherwise, when AssayFormFactor is Capillary, the option is set to be {{14 uL, 10 mg/mL},{14 uL, 8 mg/mL},{14 uL, 6 mg/mL},{14 uL, 4 mg/mL},{14 uL, 2 mg/mL}} if ReplicateDilutionCurve is True, or to be {{14 uL times NumberOfReplicates), 10 mg/mL},{14 uL times NumberOfReplicates), 8 mg/mL},{14 uL times NumberOfReplicates), 6 mg/mL},{14 uL times NumberOfReplicates), 4 mg/mL},{14 uL times NumberOfReplicates), 2 mg/mL}} if ReplicateDilutionCurve is False. When AssayFormFactor is Plate, the Sample Amount is set to a series of dilutions of 90 uL or 30 uL (depending on number of wells).
Pattern Description: List of one or more {Sample Amount, Target Concentration} entries or list of one or more {Sample Amount, Total Volume} entries or Null.
Programmatic Pattern: (({{RangeP[0*Microliter, $MaxTransferVolume], RangeP[0*Microliter, $MaxTransferVolume]}..} | {{RangeP[0*Microliter, $MaxTransferVolume], GreaterEqualP[0*(Milligram/Milliliter)] | GreaterP[0*Molar]}..}) | Automatic) | Null
Index Matches to: experiment samples
SerialDilutionCurve
The collection of dilutions that are performed on each input sample in the SampleLoadingPlate or AssayContainer. For Serial Dilution Factors, the sample will be diluted with the Buffer by the dilution factor at each transfer step. For example, a SerialDilutionCurve of {36*Microliter,{1,1.25,2,2},1} for a 100 mg/mL sample will result in 4 dilutions with concentrations of 100 mg/mL, 80 mg/mL, 40 mg/mL, and 20 mg/mL. For Serial Dilution Volumes, the Transfer Volume is taken out of the sample and added to a second well with the Buffer Volume of the Buffer. It is mixed, then the Transfer Volume is taken out of that well to be added to a third well. This is repeated to make Number Of Dilutions diluted samples. For example, if a 100 mg/mL sample with a Transfer Volume of 30 Microliters, a Buffer Volume of 10 Microliters and a Number of Dilutions of 2 is used, it will create a SerialDilutionCurve of 75 mg/mL, 56.3 mg/mL, and 42.2 mg/mL.
Default Calculation: The SerialDilutionCurve is set to Null if the AssayType is SizingPolydispersity, IsothermalStability, or MeltingCurve, or if the StandardDilutionCurve option is specified. Otherwise, when AssayFormFactor is Capillary, the option is set to be {15 uL, {1, 1.25, 1.333, 1.5, 2}, 1} if ReplicateDilutionCurve is True, or to be {(15 uL times NumberOfReplicates), {1, 1.25, 1.333, 1.5, 2}, 1} if ReplicateDilutionCurve is False. When AssayFormFactor is Plate, the Sample Volume is set to 90 uL or 30 uL (depending on number of wells). In both cases, the dilution factors with respect to the input sample are {1, 1.25, 1.333, 1.5, 2}, so a 10 mg/mL sample would be diluted to {10 mg/mL, 8 mg/mL, 6 mg/mL, 4 mg/mL, 2 mg/mL}.
Pattern Description: Serial Dilution Factor or Serial Dilution Volumes or Null.
Programmatic Pattern: (({RangeP[0*Microliter, $MaxTransferVolume], RangeP[0*Microliter, $MaxTransferVolume], RangeP[1, 48, 1]} | {RangeP[0*Microliter, $MaxTransferVolume], {GreaterEqualP[1]..}, GreaterEqualP[1, 1]}) | Automatic) | Null
Index Matches to: experiment samples
DilutionMixType
Indicates the method used to mix the SampleLoadingPlate or AssayContainer used for dilution.
Default Calculation: Automatically set to Manual when AssayType is ColloidalStability, and to Null otherwise.
Pattern Description: Pipette, Vortex, or Nutator or Null.
Programmatic Pattern: ((Pipette | Vortex | Nutator) | Automatic) | Null
Index Matches to: experiment samples
DilutionNumberOfMixes
The number of pipette out and in cycles that is used to mix the sample with the Buffer to make the DilutionCurve.
Default Calculation: Automatically set to Null if the AssayType is SizingPolydispersity, IsothermalStability, or MeltingCurve; or if either the DilutionNumberOfMixes or DilutionMixRate is specified as Null; or if MixFormFactor is set to Vortex; and to 5 otherwise.
Pattern Description: Greater than or equal to 0 and less than or equal to 20 in increments of 1 or Null.
Programmatic Pattern: (RangeP[0, 20, 1] | Automatic) | Null
Index Matches to: experiment samples
DilutionMixRate
The speed at which the dilution sample is pipetted out of and into the dilution to mix the sample with the Buffer to make the DilutionCurve.
Default Calculation: Automatically set to Null if the AssayType is SizingPolydispersity, IsothermalStability, or MeltingCurve; or if either the DilutionNumberOfMixes or DilutionMixRate is specified as Null; or if MixFormFactor is set to Vortex; and to 30 uL/second otherwise.
Pattern Description: Greater than or equal to 0.4 microliters per second and less than or equal to 250 microliters per second or Null.
Programmatic Pattern: (RangeP[0.4*(Microliter/Second), 250*(Microliter/Second)] | Automatic) | Null
Index Matches to: experiment samples
DilutionMixInstrument
The instrument used to mix the dilutions in the SampleLoadingPlate or AssayContainer used for dilution.
Default Calculation: DilutionMixInstrument is automatically set to Model[Instrument,Pipette] if DilutionMixType is set to Pipette; to Model[Instrument,Vortex] if DilutionMixType is set to Vortex; to Model[Instrument,Nutator] if DilutionMixType is set to Nutator; and to Null if AssayType is SizingPolydispersity, IsothermalStability, or MeltingCurve.
Pattern Description: An object of type or subtype Model[Instrument, Pipette], Object[Instrument, Pipette], Model[Instrument, Vortex], Object[Instrument, Vortex], Model[Instrument, Nutator], or Object[Instrument, Nutator] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Pipette], Object[Instrument, Pipette], Model[Instrument, Vortex], Object[Instrument, Vortex], Model[Instrument, Nutator], Object[Instrument, Nutator]}] | Automatic) | Null
Index Matches to: experiment samples
Sample and Solvent Information
SolventName
For each input sample, the name of the solvent specified in the DynaPro software. This option is only necessary if the AssayFormFactor is Plate and either CollectStaticLightScattering is True or the AssayType is ColloidalStability.
Default Calculation: If the AssayFormFactor is Plate and either CollectStaticLightScattering is True or the AssayType is ColloidalStability, SolventName is set based on SolventViscosity and SolventRefractiveIndex if those are specified, set to "Water" if the Buffer is "Water", or to "New" if otherwise unspecified.
Pattern Description: New or {Water, Acetic Acid 0.5%, Acetic Acid 1%, Acetic Acid 2%, Acetic Acid 3%, Acetic Acid 4%, Acetic Acid 5%, Acetic Acid 6%, Acetic Acid 7%, Acetic Acid 8%, Acetic Acid 9%, Acetone 0.5%, Acetone 1.%, Acetone 1.5%, Acetone 2.%, Acetone 2.5%, Acetone 3.%, Acetone 3.5%, Acetone 4.%, Acetone 4.5%, Acetone 5.%, Acetone 6%, Acetone 100%, Ammonium Chloride 0.5%, Ammonium Chloride 1%, Ammonium Chloride 2%, Ammonium Chloride 3%, Ammonium Chloride 4%, Ammonium Chloride 5%, Ammonium Chloride 6%, Ammonium Chloride 7%, Ammonium Chloride 8%, Ammonium Chloride 9%, Ammonium Chloride 10%, Calcium Chloride 0.5%, Calcium Chloride 1%, Calcium Chloride 2%, Calcium Chloride 3%, Calcium Chloride 4%, Calcium Chloride 5%, Calcium Chloride 6%, Calcium Chloride 7%, Calcium Chloride 8%, Calcium Chloride 9%, Calcium Chloride 10%, Cyclohexane, Ethanol 1%, Ethanol 2%, Ethanol 3%, Ethanol 4%, Ethanol 5%, Ethanol 6%, Ethanol 7%, Ethanol 8%, Ethanol 9%, Ethanol 10%, Ethanol 15%, Ethanol 20%, Ethanol 100%, Ethylene Glycol 1%, Ethylene Glycol 2%, Ethylene Glycol 3%, Ethylene Glycol 4%, Ethylene Glycol 5%, Ethylene Glycol 6%, Ethylene Glycol 7%, Ethylene Glycol 8%, Ethylene Glycol 9%, Ethylene Glycol 10%, Ethylene Glycol 20%, Glucose 1%, Glucose 2%, Glucose 3%, Glucose 4%, Glucose 5%, Glucose 6%, Glucose 7%, Glucose 8%, Glucose 9%, Glucose 10%, Glucose 20%, Glycerol 1%, Glycerol 2%, Glycerol 3%, Glycerol 4%, Glycerol 5%, Glycerol 6%, Glycerol 7%, Glycerol 8%, Glycerol 9%, Glycerol 10%, Glycerol 15%, Glycerol 20%, Glycerol 25%, Glycerol 30%, Glycerol 40%, Glycerol 50%, Glycerol 100%, Hexane, Lithium Chloride 0.5%, Lithium Chloride 1%, Lithium Chloride 2%, Lithium Chloride 3%, Lithium Chloride 4%, Lithium Chloride 5%, Lithium Chloride 6%, Lithium Chloride 7%, Lithium Chloride 8%, Lithium Chloride 9%, Lithium Chloride 10%, Magnesium Chloride 0.5%, Magnesium Chloride 1%, Magnesium Chloride 2%, Magnesium Chloride 3%, Magnesium Chloride 4%, Magnesium Chloride 5%, Magnesium Chloride 6%, Magnesium Chloride 7%, Magnesium Chloride 8%, Magnesium Chloride 9%, Magnesium Chloride 10%, Maltose 0.5%, Maltose 1%, Maltose 2%, Maltose 3%, Maltose 4%, Maltose 5%, Maltose 6%, Maltose 7%, Maltose 8%, Maltose 9%, Maltose 10%, Methanol 1%, Methanol 2%, Methanol 3%, Methanol 4%, Methanol 5%, Methanol 6%, Methanol 7%, Methanol 8%, Methanol 9%, Methanol 10%, Methanol 15%, Methanol 20%, Methanol 100%, Oleic Acid, PBS, Sodium Acetate 0.5%, Sodium Acetate 1%, Sodium Acetate 2%, Sodium Acetate 3%, Sodium Acetate 4%, Sodium Acetate 5%, Sodium Acetate 6%, Sodium Acetate 7%, Sodium Acetate 8%, Sodium Acetate 9%, Sodium Acetate 10%, Sodium Bromide 0.5%, Sodium Bromide 1%, Sodium Bromide 2%, Sodium Bromide 3%, Sodium Bromide 4%, Sodium Bromide 5%, Sodium Bromide 6%, Sodium Bromide 7%, Sodium Bromide 8%, Sodium Bromide 9%, Sodium Bromide 10%, Sodium Chloride 0.1%, Sodium Chloride 0.2%, Sodium Chloride 0.3%, Sodium Chloride 0.4%, Sodium Chloride 0.5%, Sodium Chloride 1.%, Sodium Chloride 1.5%, Sodium Chloride 2.%, Sodium Chloride 2.5%, Sodium Chloride 3.%, Sodium Chloride 3.5%, Sodium Chloride 4.%, Sodium Chloride 4.5%, Sodium Chloride 5.%, Sodium Chloride 6%, Sodium Chloride 7%, Sodium Chloride 8%, Sodium Chloride 9%, Sodium Chloride 10%, Sodium Chloride 15%, Sodium Chloride 20%, Sodium Hydroxide 0.5%, Sodium Hydroxide 1%, Sodium Hydroxide 2%, Sodium Hydroxide 3%, Sodium Hydroxide 4%, Sodium Hydroxide 5%, Sodium Hydroxide 6%, Sodium Hydroxide 7%, Sodium Hydroxide 8%, Sodium Hydroxide 9%, Sodium Hydroxide 10%, Sodium Nitrate 0.5%, Sodium Nitrate 1%, Sodium Nitrate 2%, Sodium Nitrate 3%, Sodium Nitrate 4%, Sodium Nitrate 5%, Sodium Nitrate 6%, Sodium Nitrate 7%, Sodium Nitrate 8%, Sodium Nitrate 9%, Sodium Nitrate 10%, Sodium Sulfate 0.5%, Sodium Sulfate 1%, Sodium Sulfate 2%, Sodium Sulfate 3%, Sodium Sulfate 4%, Sodium Sulfate 5%, Sodium Sulfate 6%, Sodium Sulfate 7%, Sodium Sulfate 8%, Sodium Sulfate 9%, Sodium Sulfate 10%, Sucrose 1%, Sucrose 2%, Sucrose 3%, Sucrose 4%, Sucrose 5%, Sucrose 6%, Sucrose 7%, Sucrose 8%, Sucrose 9%, Sucrose 10%, Sucrose 15%, Sucrose 20%, Toluene, Urea 1%, Urea 5%, Urea 6%, Urea 7%, Urea 8%, Urea 9%, Urea 10%, Urea 15%, Urea 20%} or Null.
Programmatic Pattern: (("New" | DLSSolventNameP) | Automatic) | Null
Index Matches to: experiment samples
SolventViscosity
For each input sample, the viscosity of the solvent specified in the DynaPro software. This option is only necessary if the AssayFormFactor is Plate and either CollectStaticLightScattering is True or the AssayType is ColloidalStability.
Default Calculation: If the AssayFormFactor is Plate and either CollectStaticLightScattering is True or the AssayType is ColloidalStability, SolventViscosity is set based on SolventName if it is specified or resolved, as described in $DLSSolventLookupTable, and set to Null otherwise.
Pattern Description: Greater than or equal to 0 centipoise and less than or equal to 50 centipoise or Null.
Programmatic Pattern: (RangeP[0*Centipoise, 50*Centipoise] | Automatic) | Null
Index Matches to: experiment samples
SolventRefractiveIndex
For each input sample, the viscosity of the solvent specified in the DynaPro software. This option is only necessary if the AssayFormFactor is Plate and either CollectStaticLightScattering is True or the AssayType is ColloidalStability.
Default Calculation: If the AssayFormFactor is Plate and either CollectStaticLightScattering is True or the AssayType is ColloidalStability, SolventRefractiveIndex is set based on SolventName if it is specified or resolved, as described in $DLSSolventLookupTable, and set to Null otherwise.
Pattern Description: Greater than or equal to 1 and less than or equal to 3 or Null.
Programmatic Pattern: (RangeP[1, 3] | Automatic) | Null
Index Matches to: experiment samples
Analyte
For each input sample, the Model[Molecule] member of the Composition field whose concentration is used to collect static light scattering data, or to calculate B22/A2 and kD or G22 when the AssayType is ColloidalStability.
Default Calculation: Automatically set to Null when CollectStaticLightScattering is False or AssayFormFactor is Capillary and the AssayType is not ColloidalStability. When AssayType is ColloidalStability, or when CollectStaticLightScattering is True, automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule] or Null.
Programmatic Pattern: (ObjectP[Model[Molecule]] | Automatic) | Null
Index Matches to: experiment samples
AnalyteMassConcentration
For each input sample, the initial mass concentration of the Analyte before any dilutions outlined by the StandardDilutionCurve or SerialDilutionCurve are performed when the AssayType is ColloidalStability.
Default Calculation: Automatically set to Null when CollectStaticLightScattering is False or AssayFormFactor is Capillary and the AssayType is not ColloidalStability. When AssayType is ColloidalStability, or when CollectStaticLightScattering is True, automatically set to the mass concentration of the first value in the Analytes field of the input sample, or, if not populated, to that of the first analyte in the Composition field of the input sample, or if none exist, that of the first identity model of any kind in the Composition field.
Pattern Description: Greater than or equal to 0 milligrams per milliliter or greater than 0 molar or Null.
Programmatic Pattern: ((GreaterEqualP[0*(Milligram/Milliliter)] | GreaterP[0*Molar]) | Automatic) | Null
Index Matches to: experiment samples
AnalyteRefractiveIndexIncrement
For each input sample, the known or estimated refractive index increment (also known as dn/dc) of the Analyte.
Default Calculation: Automatically set to Null when CollectStaticLightScattering is False or AssayFormFactor is Capillary and the AssayType is not ColloidalStability. When AssayType is ColloidalStability, or when CollectStaticLightScattering is True and AssayFormFactor is Plate, automatically set to 0.185 Milliliter/Gram if the Analyte is a Model[Molecule, Protein] or Model[Molecule, cDNA], and to 0.15 Milliliter/Gram if the Analyte is a Model[Molecule, Polymer] or Model[Molecule, Oligomer].
Pattern Description: Greater than or equal to 0 milliliters per gram or Null.
Programmatic Pattern: (GreaterEqualP[0*(Milliliter/Gram)] | Automatic) | Null
Index Matches to: experiment samples
Buffer
The buffer or solvent that is used to dilute the sample to make a DilutionCurve or in collecting static light scattering.
Default Calculation: The Buffer is automatically set to Null when CollectStaticLightScattering is False or AssayFormFactor is Capillary and the AssayType is not ColloidalStability. When AssayType is ColloidalStability, or when CollectStaticLightScattering is True, the Buffer is set to the BlankBuffer if it is specified, to the Solvent of the input sample if the BlankBuffer is not specified, and to Model[Sample, "Milli-Q water"] otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
BlankBuffer
The sample that is used as a 0 mg/mL blank in ColloidalStability assays or when CollectStaticLightScattering is True, to determine the diffusion coefficient at infinite dilution.
Default Calculation: The BlankBuffer is set to the Buffer if the AssayType is ColloidalStability or if CollectStaticLightScattering is True and to Null otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
Colloidal Stability
CalculateMolecularWeight
When AssayFormFactor is Plate, determines if Static Light Scattering (SLS) is used to calculate weight-average molecular weight.
Default Calculation: When AssayFormFactor is Plate and AssayType is ColloidalStability, automatically set to True. when CollectStaticLightScattering is False or AssayFormFactor is Capillary or AssayFormFactor is Capillary, automatically set to Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Light Scattering
CollectStaticLightScattering
Indicates whether static light scattering (SLS) data are collected along with DLS data. Reliable static light scattering data can be obtained only if the combination of plate and solvent has previously been calibrated.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
CalibratePlate
When AssayFormFactor is Plate, indicates whether the AssayContainer should be calibrated by measuring the scattered light intensities of a series of dilutions of a standard sample before any other data collection.
Default Calculation: When AssayFormFactor is Plate and CollectStaticLightScattering is True, automatically set to True.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
NumberOfAcquisitions
For each Dynamic Light Scattering (DLS) measurement, the number of series of speckle patterns that are each collected over the AcquisitionTime to create the measurement's autocorrelation curve.
Pattern Description: Greater than or equal to 1 and less than or equal to 20 in increments of 1.
Programmatic Pattern: RangeP[1, 20, 1]
AcquisitionTime
For each Dynamic Light Scattering (DLS) measurement, the length of time that each acquisition generates speckle patterns to create the measurement's autocorrelation curve.
Pattern Description: Greater than or equal to 1 second and less than or equal to 30 seconds.
Programmatic Pattern: RangeP[1*Second, 30*Second]
AutomaticLaserSettings
Indicates if the LaserPower and DiodeAttenuation are automatically set at the beginning of the assay by the Instrument to levels ideal for the samples, such that the count rate falls within an optimal, predetermined range.
Default Calculation: Automatically set to False if either LaserPower or DiodeAttenuation is not Null, and to True otherwise.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
LaserPower
The Percent of the max laser power that is used to make Dynamic Light Scattering (DLS) measurements. The laser level is optimized at run time by the instrument software when AutomaticLaserSettings is True and LaserLevel is Null.
Default Calculation: The LaserPower option is set to 100Percent if AutomaticLaserSettings is False and to Null otherwise.
Pattern Description: Greater than or equal to 1 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[1*Percent, 100*Percent] | Automatic) | Null
DiodeAttenuation
The Percent of scattered signal that is allowed to reach the avalanche photodiode (in Capillary-type assays) or prevented from reaching the avalanche photodiode (in Plate-type assays). The attenuator level is optimized at run time by the instrument software when AutomaticLaserSettings is True and DiodeAttenuation is Null.
Default Calculation: If AutomaticLaserSettings is False and AssayFormFactor is Plate, the DiodeAttenuation option is set to 0Percent; if AutomaticLaserSettings is False and AssayFormFactor is Capillary, the DiodeAttenuation option is set to 100Percent, and to Null in all other cases.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Isothermal Stability
MeasurementDelayTime
The length of time between the consecutive Dynamic Light Scattering (DLS) measurements for a specific AssayContainer well during an IsothermalStability assay. The duration of the experiment is set either by this option or by the total IsothermalRunTime.
Default Calculation: When AssayType is IsothermalStability and when AssayFormFactor is Capillary, the MeasurementDelayTime option is set to a value calculated by the Instrument manufacture's proprietary algorithm (dependent on the number of input samples and the IsothermalAttenuatorAdjustment); when AssayFormFactor is Plate, MeasurementDelayTime is automatically set to 1 hour. When AssayType is SizingPolydispersity, MeltingCurve, or ColloidalStability, automatically set to Null.
Pattern Description: Greater than or equal to 70 seconds and less than or equal to 3.75 hours or Null.
Programmatic Pattern: (RangeP[70*Second, 3.75*Hour] | Automatic) | Null
IsothermalMeasurements
The number of separate DLS measurements that are made during the IsothermalStability assay, either separated by MeasurementDelayTime or distributed over IsothermalRunTime.
Default Calculation: The IsothermalMeasurements option is set to 10 if the AssayType is IsothermalStability and the IsothermalRunTime is Null, or to Null otherwise.
Pattern Description: Greater than or equal to 0 and less than or equal to 8022 in increments of 1 or Null.
Programmatic Pattern: (RangeP[0, 8022, 1] | Automatic) | Null
IsothermalRunTime
The total length of the IsothermalStability assay during which the IsothermalMeasurements number of Dynamic Light Scattering (DLS) measurements are made. The duration of the experiment is set either by this option or by the MeasurementDelayTime.
Default Calculation: The IsothermalRunTime is set to 10 times the MeasurementDelayTime if the AssayType is IsothermalStability and the IsothermalMeasurements option is Null, or to Null otherwise.
Pattern Description: Greater than or equal to 70 seconds and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[70*Second, $MaxExperimentTime] | Automatic) | Null
IsothermalAttenuatorAdjustment
Indicates if the attenuator level is automatically set for each DLS measurement throughout the IsothermalStability assay. If First, the attenuator level is automatically set for the first DLS measurement and the same level is used throughout the assay.
Default Calculation: The IsothermalAttenuatorAdjustment is set to Every if the AssayType is IsothermalStability and to Null otherwise.
Pattern Description: First or Every or Null.
Programmatic Pattern: ((First | Every) | Automatic) | Null
Melting Curve
MinTemperature
The low temperature of the heating or cooling curve; the starting temperature when TemperatureRampOrder is {Heating,Cooling}.
Default Calculation: When AssayType is MeltingCurve, automatically set to 4 Celsius if AssayFormFactor is Plate and to 15 Celsius when AssayFormFactor is Capillary. When AssayType is SizingPolydispersity, IsothermalStability, or ColloidalStability, automatically set to Null.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[4*Celsius, 100*Celsius] | Automatic) | Null
MaxTemperature
The high temperature of the heating or cooling curve; the starting temperature when TemperatureRampOrder is {Cooling,Heating}.
Default Calculation: When AssayType is MeltingCurve, automatically set to 85 Celsius if AssayFormFactor is Plate and 95 Celsius if AssayFormFactor is Capillary. When AssayType is SizingPolydispersity, IsothermalStability, or ColloidalStability, automatically set to Null.
Pattern Description: Greater than or equal to 4 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[4*Celsius, 100*Celsius] | Automatic) | Null
TemperatureRampOrder
The order of temperature ramping (i.e., heating followed by cooling or vice versa) to be performed in each cycle. Heating is defined as going from MinTemperature to MaxTemperature; cooling is defined as going from MaxTemperature to MinTemperature.
Default Calculation: When AssayType is MeltingCurve, automatically set to {Heating,Cooling}. When AssayType is SizingPolydispersity, IsothermalStability, or ColloidalStability, automatically set to Null.
Pattern Description: {Heating, Cooling} or {Cooling, Heating} or Null.
Programmatic Pattern: (ThermodynamicCycleP | Automatic) | Null
NumberOfCycles
The number of instances of repeated heating and cooling (or vice versa) cycles.
Default Calculation: When AssayType is MeltingCurve, automatically set to 1 cycle. When AssayType is SizingPolydispersity, IsothermalStability, or ColloidalStability, automatically set to Null.
Pattern Description: 0.5, 1, 2, or 3 or Null.
Programmatic Pattern: ((0.5 | 1 | 2 | 3) | Automatic) | Null
TemperatureRamping
The type of temperature ramp. Linear temperature ramps increase temperature at a constant rate given by TemperatureRampRate. Step temperature ramps increase the temperature by TemperatureRampStep/TemperatureRampStepTime and holds the temperature constant for TemperatureRampStepHold before measurement.
Default Calculation: When AssayType is MeltingCurve, automatically set to Step if NumberOfTemperatureRampSteps or StepHoldTime are specified and to Linear if they are not specified. When AssayType is SizingPolydispersity, IsothermalStability, or ColloidalStability, automatically set to Null.
Pattern Description: Linear or Step or Null.
Programmatic Pattern: ((Linear | Step) | Automatic) | Null
TemperatureRampRate
The absolute value of the rate at which the temperature is changed in the course of one heating/cooling cycle.
Default Calculation: When AssayType is MeltingCurve, if TemperatureRamping is Linear, automatically set to 1 Celsius/Minute (when AssayFormFactor is Capillary) or 0.25 Celsius/Minute (when AssayFormFactor is Plate), and if TemperatureRamping is Step, automatically set to the max ramp rate available on the instrument.
Pattern Description: Greater than or equal to 0.0015 degrees Celsius per second and less than or equal to 3.4 degrees Celsius per second or Null.
Programmatic Pattern: (RangeP[0.0015*(Celsius/Second), 3.4*(Celsius/Second)] | Automatic) | Null
TemperatureResolution
The amount by which the temperature is changed between each data point and the subsequent data point for a given sample during the heating/cooling curves.
Default Calculation: When AssayType is MeltingCurve, automatically set to highest possible resolution available for the Instrument. When AssayType is SizingPolydispersity, IsothermalStability, or ColloidalStability, automatically set to Null.
Pattern Description: Greater than or equal to 0 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[0*Celsius, 100*Celsius] | Automatic) | Null
NumberOfTemperatureRampSteps
The number of step changes in temperature for a heating or cooling cycle.
Default Calculation: When AssayType is MeltingCurve, if TemperatureRamping is Step, automatically set to the absolute value of the difference in MinTemperature and MaxTemperature divided by 1 Celsius, rounded to the nearest integer; if TemperatureRamping is Linear, automatically set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 100 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 100, 1] | Automatic) | Null
StepHoldTime
The length of time samples are held at each temperature during a stepped temperature ramp prior to light scattering measurement.
Default Calculation: When AssayType is MeltingCurve, automatically set to 30 Second when TemperatureRamping is set to Step and Null when TemperatureRamping is set to Linear.
Pattern Description: Greater than or equal to 1 second and less than or equal to 3.75 hours or Null.
Programmatic Pattern: (RangeP[1*Second, 3.75*Hour] | Automatic) | Null
Sample Storage
SampleLoadingPlateStorageCondition
The conditions under which any leftover samples from the StandardDilutionCurve or SerialDilutionCurve are stored in the SampleLoadingPlate after the samples are transferred to the AssayContainer(s).
Default Calculation: Automatically set to Null when AssayFormFactor is Plate and to Disposal when AssayFormFactor is Capillary.
Pattern Description: {{AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal} or an object of type or subtype Model[StorageCondition] or Null.
Programmatic Pattern: ((ObjectP[Model[StorageCondition]] | (SampleStorageTypeP | Disposal)) | Automatic) | Null
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed.
Pattern Description: {{AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal} or an object of type or subtype Model[StorageCondition] or Null.
Programmatic Pattern: ((ObjectP[Model[StorageCondition]] | (SampleStorageTypeP | Disposal)) | Automatic) | Null
Index Matches to: experiment samples
SamplesOutStorageCondition
The non-default conditions under which the SamplesOut of this experiment should be stored after the protocol is completed.
Default Calculation: When AssayFormFactor is Capillary, automatically set to Disposal. When AssayFormFactor is Plate, if all of the experiment samples have the same StorageCondition, automatically set to that StorageCondition; if the experiment samples have different StorageConditions, automatically set to Disposal; if no StorageConditions are available for the experiment samples, automatically set to AmbientStorage.
Pattern Description: {{AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal} or an object of type or subtype Model[StorageCondition] or Null.
Programmatic Pattern: ((ObjectP[Model[StorageCondition]] | (SampleStorageTypeP | Disposal)) | Automatic) | Null
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and when the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise set to Model[Container, Vessel, "2mL Tube"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[Model[Container]] | Automatic) | Null
Index Matches to: experiment samples
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
Index Matches to: experiment samples