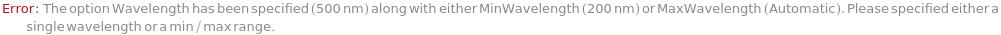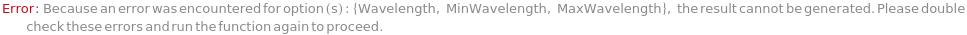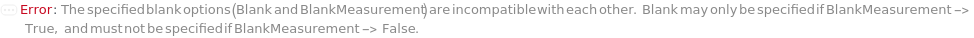ExperimentUVMelting
ExperimentUVMelting[Samples]⟹Protocol
generates a Protocol object for assessing the dissociation characteristics on the provided Samples during heating and cooling (melting curve analysis) using UV absorbance measurements.
ExperimentUVMelting measures the UV absorbance of hybridized nucleic acids across a temperature gradient. Double-stranded nucleic acids like DNA, display an ultraviolet absorption peak at 260 nm which increases upon thermal dissociation of the molecule into complementary single strands, also known as "melting" or "denaturation". In this experiment, absorbance can be monitored during a heating temperature ramp where strands are denatured and a cooling temperature ramp where strands are hybridized. The data generated by this experiment can be used to determine the sample melting point which is characteristic of the sample sequence, length, concentration, sequence mismatch, and solvent conditions.
Experimental Principles
Figure 1.1: Step1: Samples can be incubated, centrifuged, filtered, and/or aliquoted in a compatible container using the shared sample preparation framework. Step2: Prepared samples are transferred and pooled as desired into instrument compatible cuvettes. Samples can then be mixed and incubated using pooled sample options. Step3: Prepared samples are equilibrated to the starting temperature of the assay. Step4: Samples are repeatedly excited by the xenon flash lamp and UV absorbance is read simultaneously for all cuvettes during the temperature ramp. Acquisition intervals are determined by TemperatureResolution option. Steps 3 & 4 are repeated for each heating and cooling cycle. Step5: Samples are tranfered out of cuvettes for storage. Step6: Data is assessed using AnalyzeMeltingPoint to determine the melting temperature of samples.
Instrumentation
Cary 3500
Figure 2.1: The Cary 3500 is a UV/Vis spectrometer with the capacity to interrogate seven samples and one reference sample simultaneously. The temperature for each pair of sample cuvettes is controlled by Peltier modules for a total of 4 independent temperature zones that can range from -10 Celsius to 100 Celsius. Currently ExperimentUVMelting supports a single temperature profile for all temperature zones. Thermal profiles can be programmed into the instrument to interrogate samples at multiple temperatures. Sample temperature can either be recorded from the cuvette block temperature or by immersion probes. The light source is a xenon flash lamp. The light beam from the lamp is split into different wavelengths ranging from 190 nm to 900 nm and the desired wavelength is isolated using a monochromator. The incident light beam then passes into the sample module where it is split into 8 beams by a fiber optic beam splitter. Each beam excites one sample cuvette and the emitted light is then collected by the instrument detector.
Experiment Options
Method
Instrument
Pattern Description: An object of type or subtype Model[Instrument, Spectrophotometer] or Object[Instrument, Spectrophotometer]
Programmatic Pattern: ObjectP[{Model[Instrument, Spectrophotometer], Object[Instrument, Spectrophotometer]}] | Automatic
MaxTemperature
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 100 degrees Celsius.
MinTemperature
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
NumberOfCycles
EquilibrationTime
TemperatureRampOrder
TemperatureResolution
The amount by which the temperature will be changed between each data point and the next during the melting and/or cooling curves.
Pattern Description: Greater than or equal to 1 degree Celsius and less than or equal to 100 degrees Celsius.
TemperatureRampRate
Pattern Description: Greater than or equal to 0.1 degrees Celsius per minute and less than or equal to 40 degrees Celsius per minute.
Wavelength
Wavelength of light which should be passed through the cuvette holding the sample in order to measure the sample's absorbance.
Default Calculation: Automatically resolves to Null when MinWavelength or MaxWavelength are set, otherwise resolves to 260 nm for oligomer (DNA/RNA/PNA) samples or to 280 nm for protein samples.
Pattern Description: Greater than or equal to 190 nanometers and less than or equal to 900 nanometers or Null.
MinWavelength
The lower limit of the wavelength range of light that should be passed through the cuvette holding the sample in order to measure the sample's absorbance. Note that currently the Cary 3500 can only collect 10 wavelengths per scan, and so when specifying a wavelength range, the wavelengths actually measured will be a selection in this range that always includes the MinWavelength, the MaxWavelength, and, if included in the range, 260 Nanometer and 280 Nanometer.
Default Calculation: Automatically resolves to the highest possible setting (900 Nanometer) if MinWavelength is specified, otherwise resolves to Null.
Pattern Description: Greater than or equal to 190 nanometers and less than or equal to 900 nanometers or Null.
MaxWavelength
The upper limit of the wavelength range of light that should be passed through the cuvette holding the sample in order to measure the sample's absorbance. Note that currently the Cary 3500 can only collect 10 wavelengths per scan, and so when specifying a wavelength range, the wavelengths actually measured will be a selection in this range that always includes the MinWavelength, the MaxWavelength, and, if included in the range, 260 Nanometer and 280 Nanometer.
Default Calculation: Automatically resolves to the lowest possible wavelength (190 Nanometer) if MaxWavelength is specified, otherwise resolves to Null.
Pattern Description: Greater than or equal to 190 nanometers and less than or equal to 900 nanometers or Null.
TemperatureMonitor
BlankMeasurement
Indicates if for each pooled sample, a corresponding blank measurement with buffer only will be performed, prior to the absorbance thermodynamics run of the pooled sample.
Blank
The source used to generate a blank sample whose absorbance is subtracted as background from the absorbance readings of the input sample.
Default Calculation: Automatically set to Null if BlankMeasurement is False, or to the value of AssayVolume if that was specified and BlankMeasurement is True, or Model[Sample, "Milli-Q water"] otherwise.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
NumberOfReplicates
The number of times to repeat the experiment on each sample or list of samples, if provided. Note that if Aliquot -> True, this indicates the number of times each provided sample will be aliquoted/pooled into a cuvette before each pool is read once.
Sample Preparation
NestedIndexMatchingMix
Default Calculation: Automatically set based on whether pooling and/or dilution of the source samples will be performed.
NestedIndexMatchingMixType
Default Calculation: Automatically set to Pipette if the cuvette aperture and available tip types allow for mixing by pipette, otherwise set to Invert.
NestedIndexMatchingNumberOfMixes
Pattern Description: Greater than or equal to 1 and less than or equal to 50 in increments of 1 or Null.
NestedIndexMatchingMixVolume
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 50 milliliters or Null.
NestedIndexMatchingIncubate
PooledIncubationTime
NestedIndexMatchingIncubationTemperature
Pattern Description: Ambient or greater than or equal to 22 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
NestedIndexMatchingAnnealingTime
Duration for which the pooled samples remain in the thermal incubation instrument before being removed, allowing the system to settle to room temperature after the PooledIncubationTime has passed.
Sample Storage
ContainerOut
The desired container generated samples should be transferred into by the end of the experiment, with indices indicating grouping of samples in the same plates.
Default Calculation: Automatically set as the PreferredContainer for the total volume of the pooled samples.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Model Input
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Programmatic Pattern: ((Null | (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All)) | Automatic) | Null
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise, automatically set to Model[Container, Cuvette, "Micro Scale Black Walled UV Quartz Cuvette"].
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSamplePreparation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentratedBufferDilutionFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Warnings and Errors
Messages (32)
ContainerOutStorageConditionConflict (2)
ContainerOverOccupied (1)
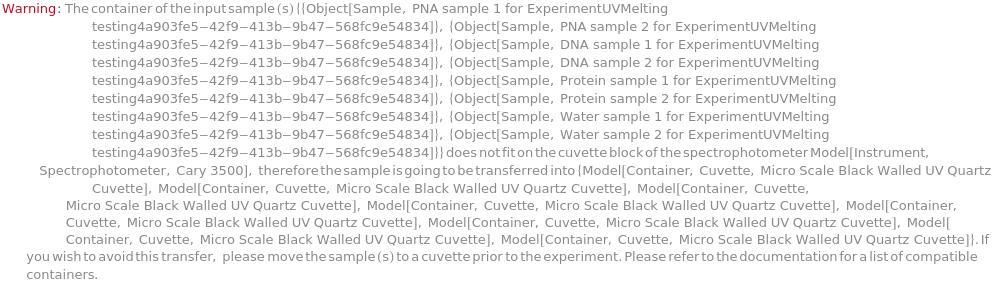
IncompatibleCuvette (1)
IncompatibleSpectrophotometer (1)
InstrumentPrecision (1)
MismatchingNestedIndexMatchingdMixOptions (1)
MismatchingNestedIndexMatchingIncubateOptions (1)
MismatchingWavelengthOptions (1)
NestedIndexMatchingMixVolumeNotCompatibleWithMixType (2)
NestedIndexMatchingVolumeBelowMinThreshold (1)
NestedIndexMatchingVolumeOutOfRange (2)
Samples with assay volume (total volume of samples and buffers inside a particular pool) smaller than the minimum working range of the smallest cuvette cannot be reliably measured:


The assay volume (total volume of samples and buffers inside a particular pool) has to fall within the working range of an available cuvette:


NoMixingDespiteAliquotting (1)
NumberOfReplicatesRequiresAliquoting (1)
ObjectDoesNotExist (4)
SamplesOutStorageConditionConflict (1)
TooManyUVMeltingSamples (1)
TransferRequired (1)
UnableToBlank (1)
Possible Issues
High ramp rate
When the ramp rate is too high, the recorded temperature from the cuvette block may not reflect the actual temperature of the samples inside the cuvettes.
Constant absorbance across temperature range
If the sample does not have structure-dependent changes in absorbance at the specified wavelength or within the wavelength range, the measured absorbance may be constant across a large temperature.
Unable to determine melting point
Last modified on Wed 17 Sep 2025 16:55:27