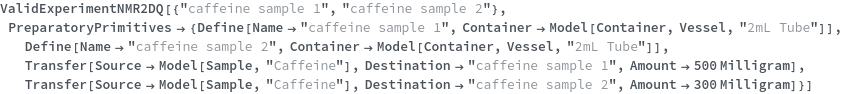
ValidExperimentNMR2DQ
ValidExperimentNMR2DQ[samples]⟹boolean
returns a boolean indicating the validity of an ExperimentNMR2D call for measuring the two-dimensional nuclear magnetic resonance (NMR) spectrum of provided samples.
Details
- This function runs a series of tests to ensure that the provided inputs/options, when passed to ExperimentNMR2D proper, will return a valid experiment.
Input

Output

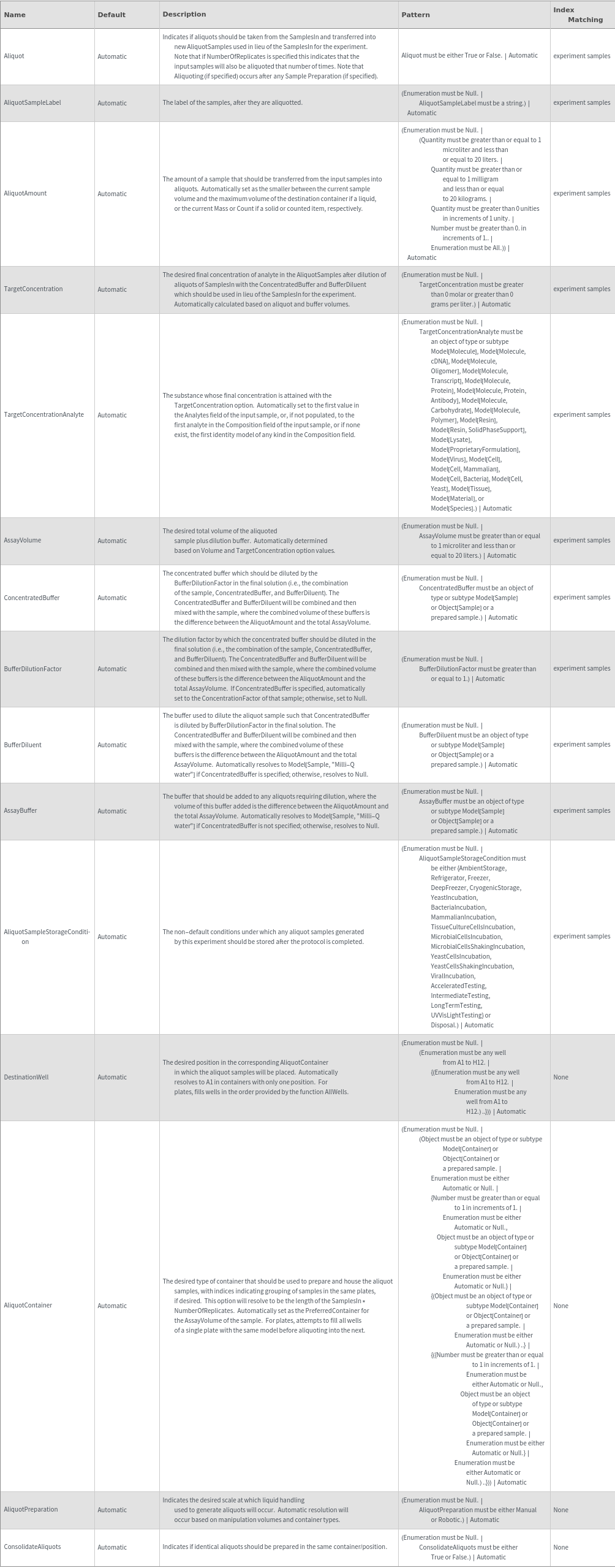
Aliquoting Options
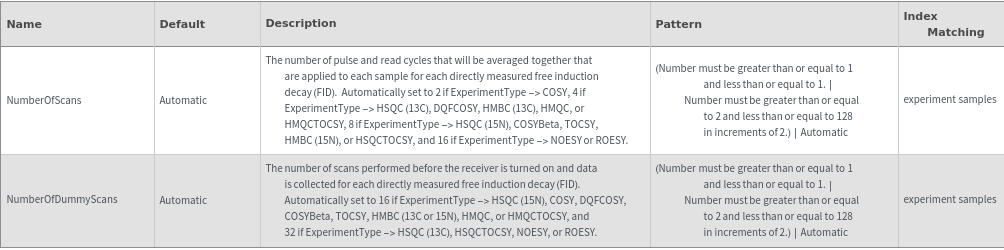
Data Acquisition Options
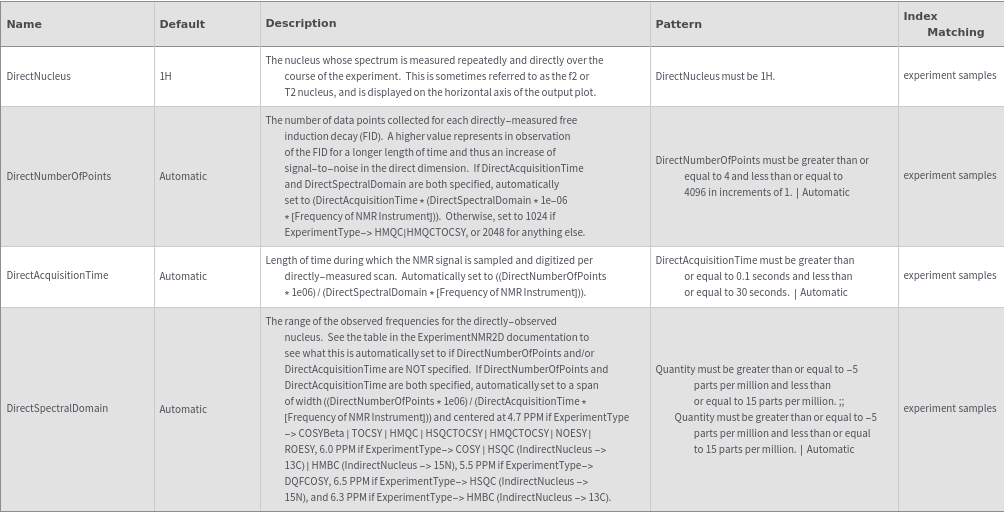
Direct Nucleus Stimulation Options
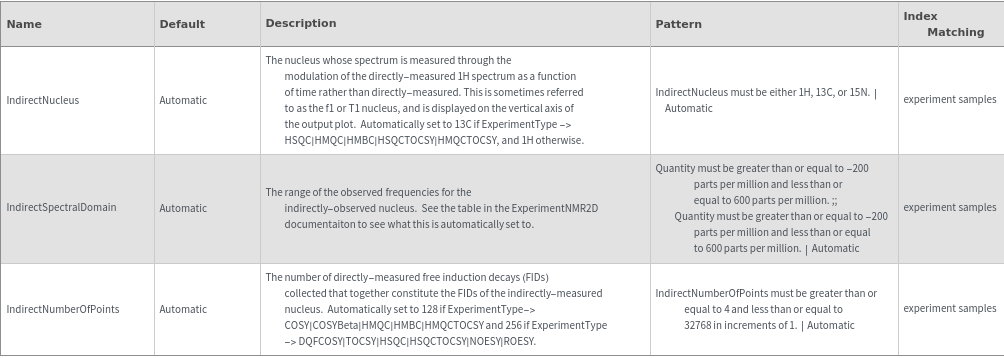
Indirect Nucleus Stimulation Options
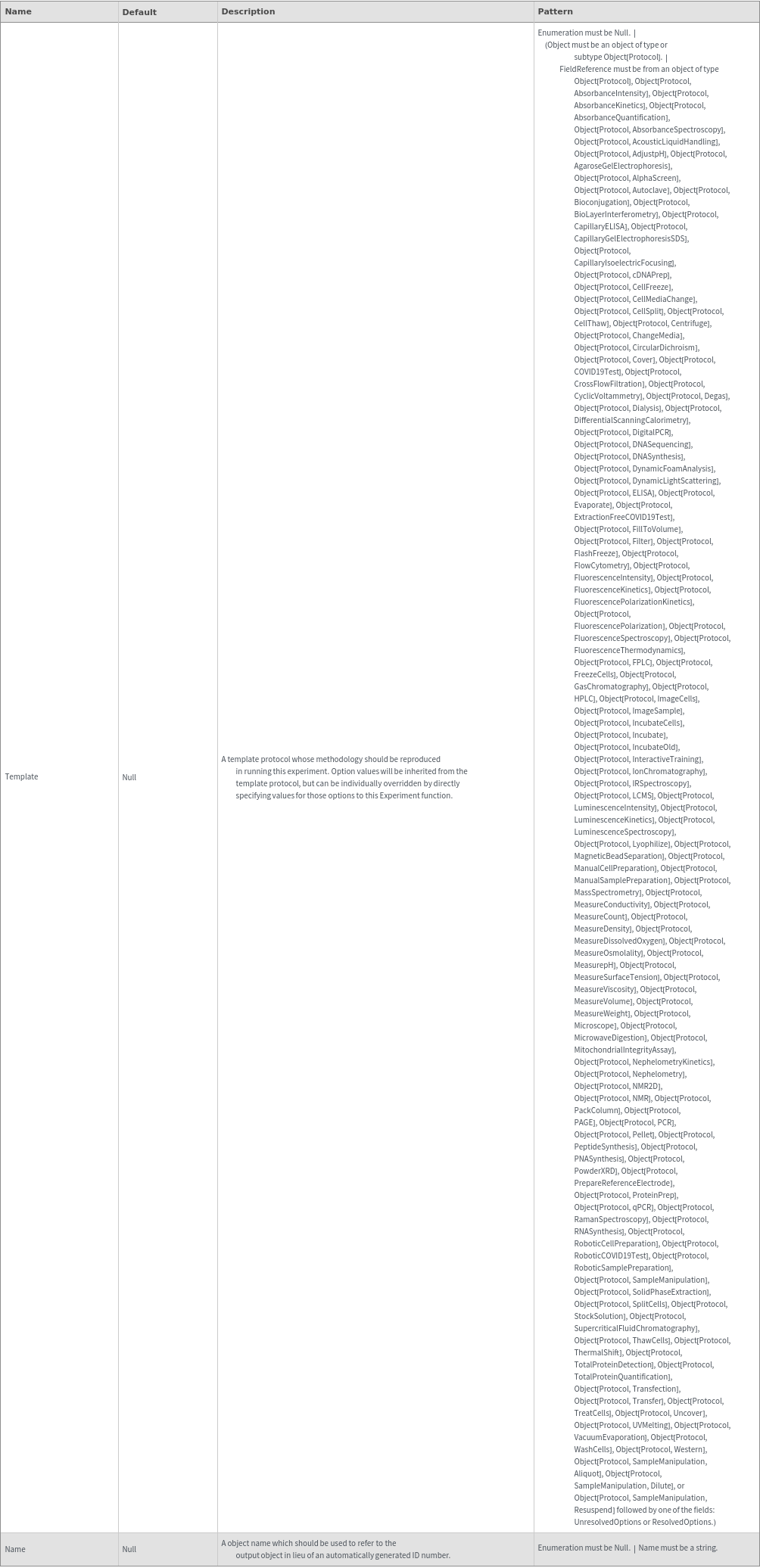
Organizational Information Options
Post Experiment Options
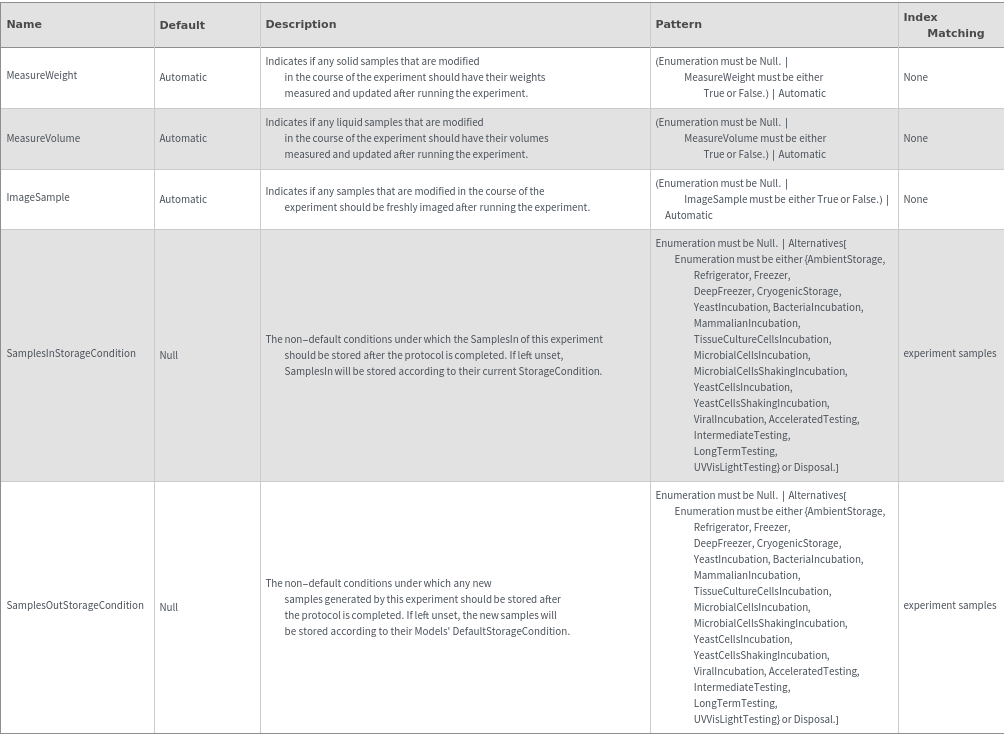
Preparatory Centrifugation Options
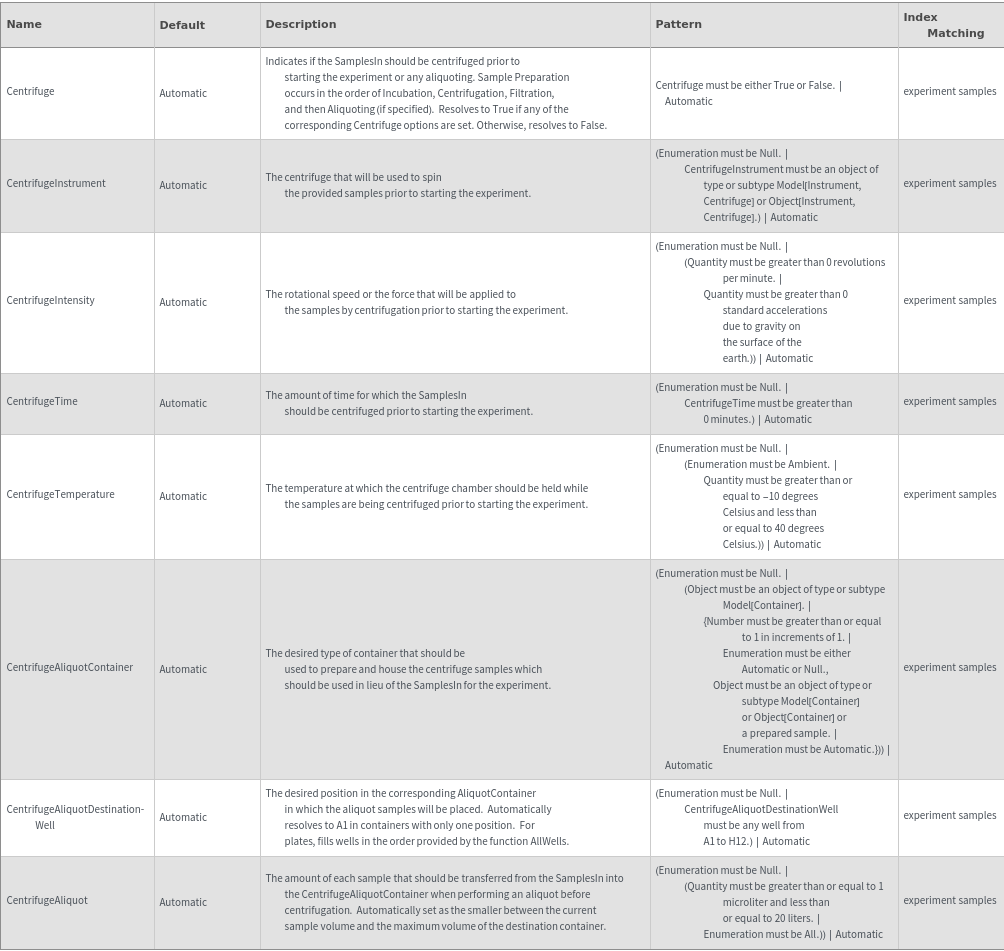
Preparatory Filtering Options
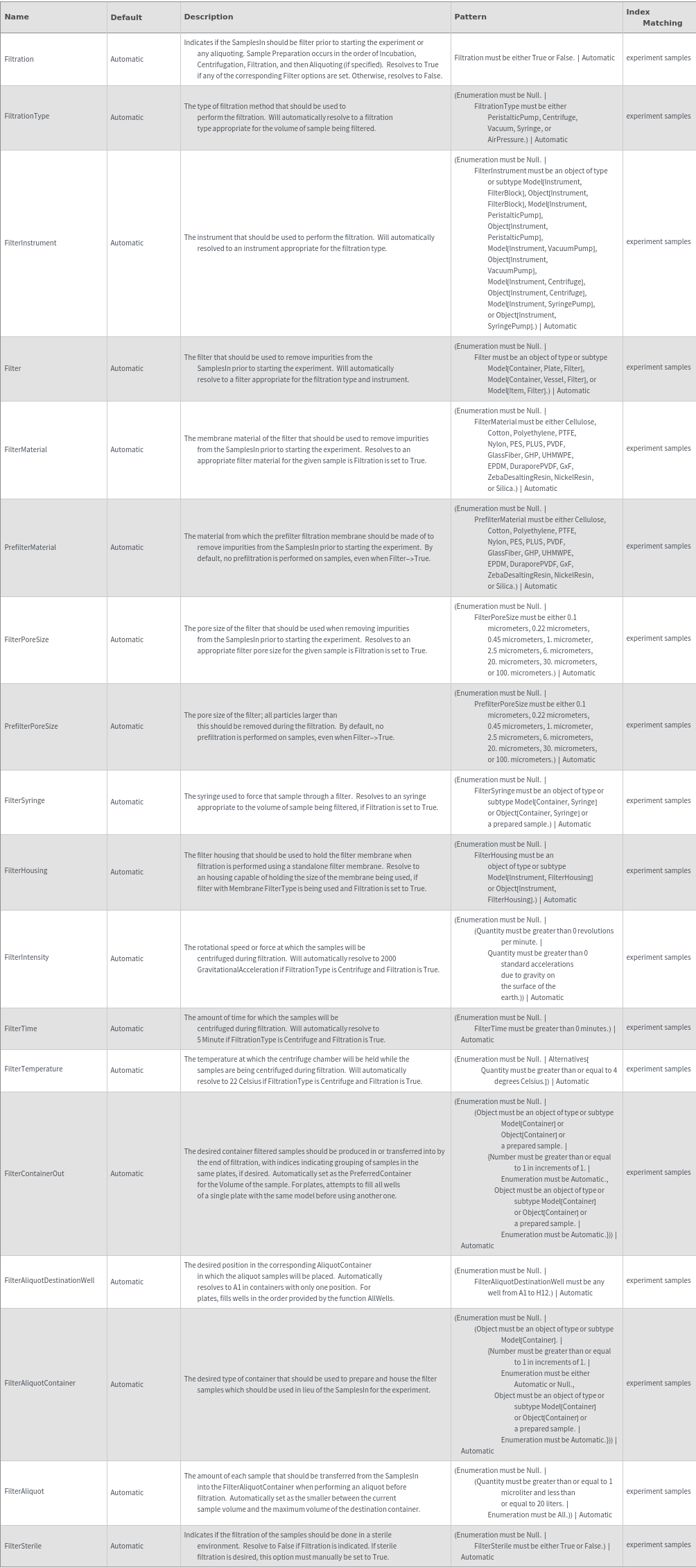
Preparatory Incubation Options
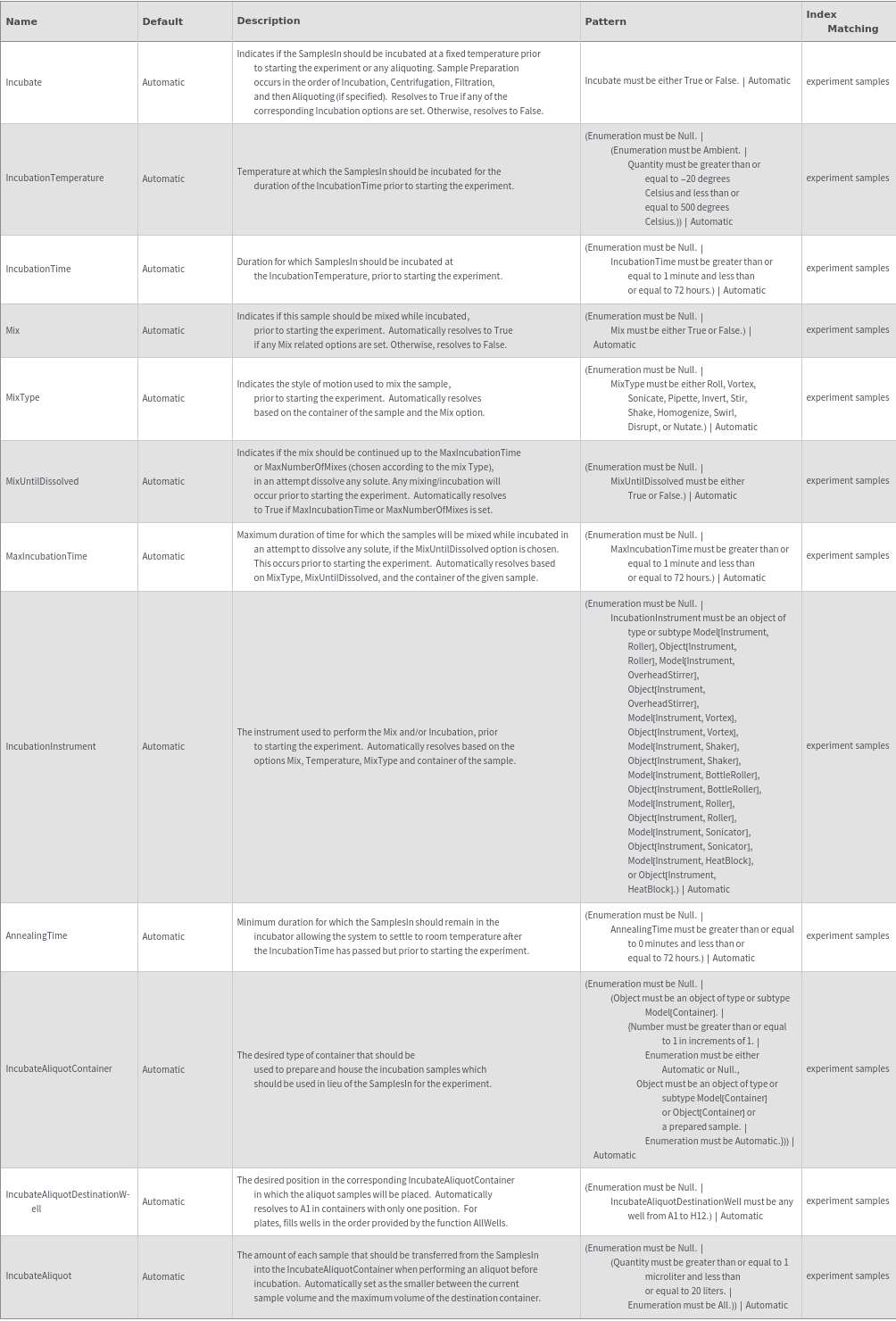
Sample Parameters Options
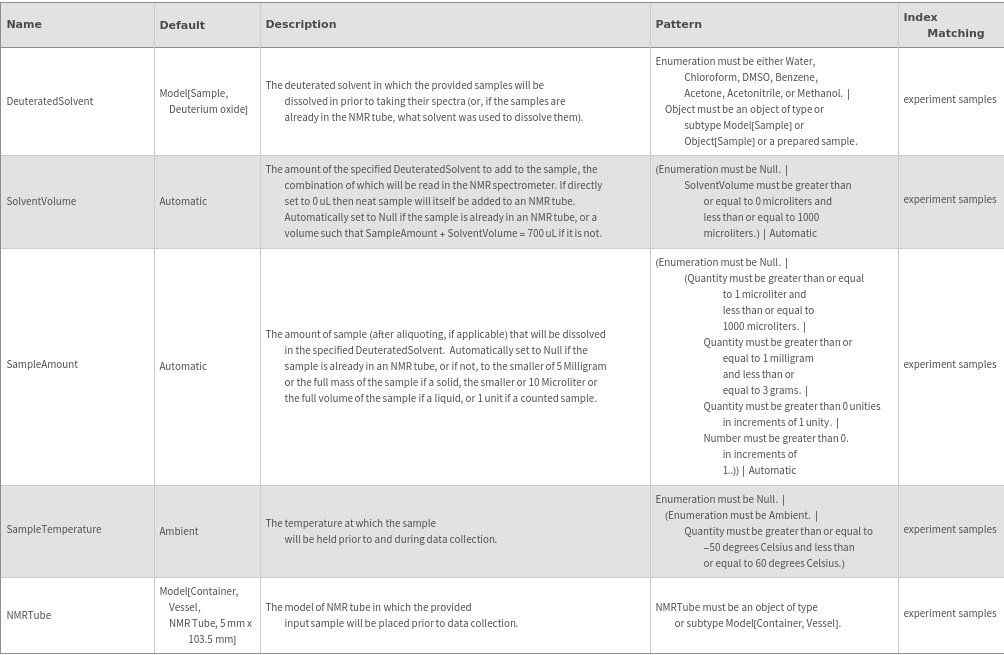
Sample Preparation Options

General Options
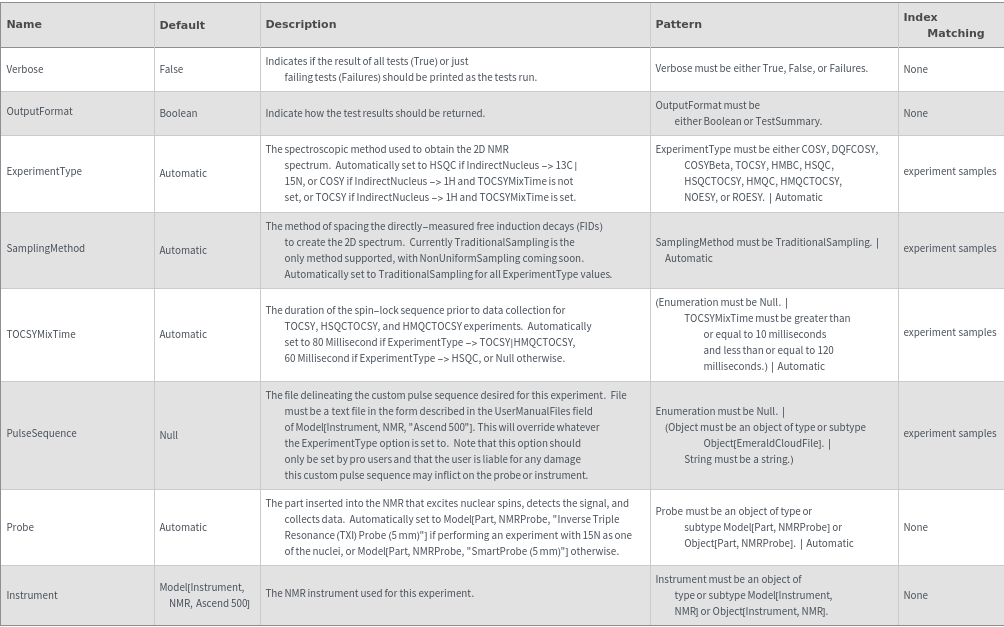
Protocol Options