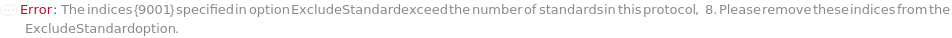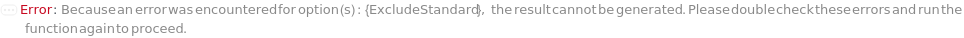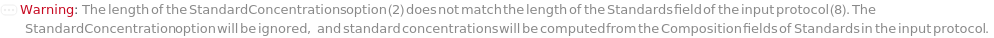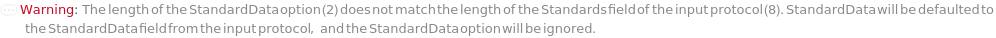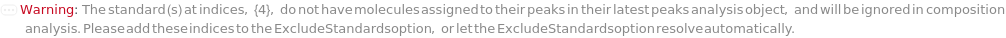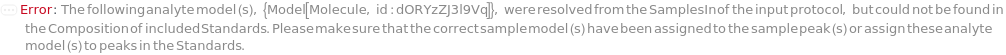AnalyzeComposition
AnalyzeComposition[Protocol]⟹Object
analyzes the chemical composition of chromatogram peaks in a HPLC protocol.
Details
Input

Output

Standards Options

Upload Options

Method Options

Examples
open allclose allBasic Examples (3)
Concentrations are calculated by fitting, for each analyte, a standard curve mapping analyte peak area to analyte concentration in input protocol Standards. Both standards and assays must have their HPLC data peak-picked. Analytes are considered to be the set of all Model[Molecule] objects assigned to peaks in the StandardData objects:
Given an HPLC protocol containing Standards with known concentrations of analytes, and SamplesIn containing unknown assays, compute the concentration of analytes in each assay sample:
The concentrations of standards and assays are stored in the StandardCompositions and AssayCompositions fields of the resulting analysis object:
Options (9)
PreferredConcentrationType (2)
AnalyzeComposition will attempt to convert concentrations to MolarConcentration by default:
Attempt to convert concentrations to mass concentrations. If not enough information (molecular weights, densities, sample volumes) is available from the analyte models, concentrations will remain in their original units:
StandardConcentrations (2)
Explicitly specify the concentrations of analyte for each Standard object in the protocol with arbitrary concentration units, using a list of {analyte,concentration} pairs for each Standard:
StandardConcentrations will default to concentrations computed from the Composition field(s) of the objects in the Standard field of input protocol. Empty entries {} indicate no analytes could be assigned to picked-peaks in StandardData: