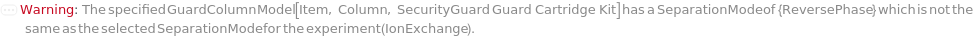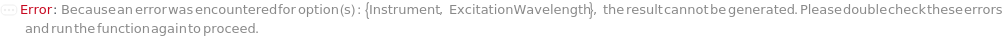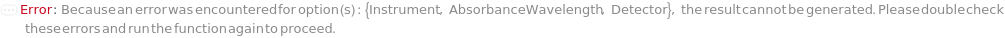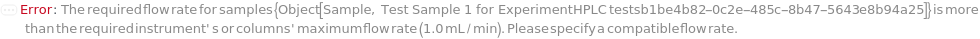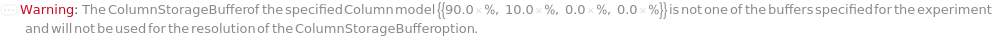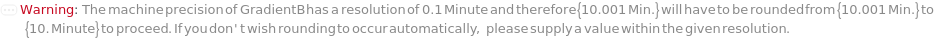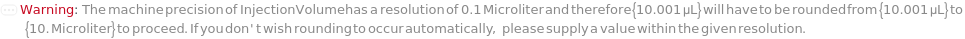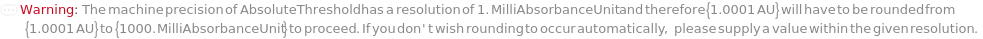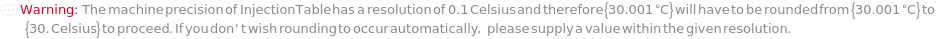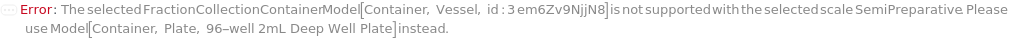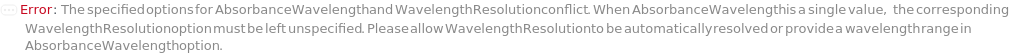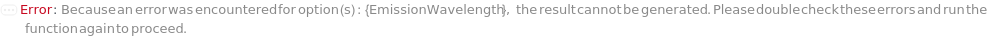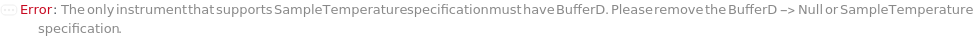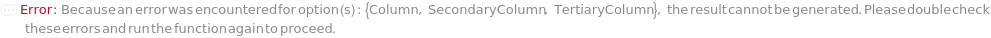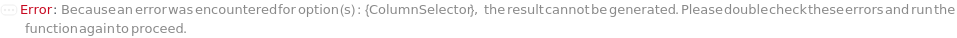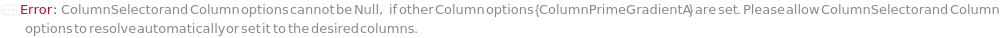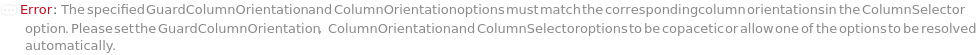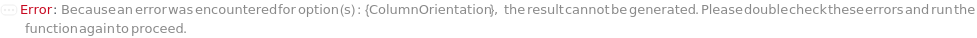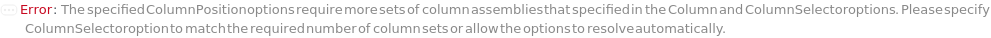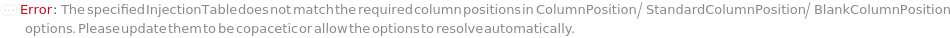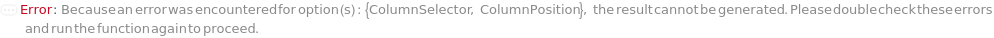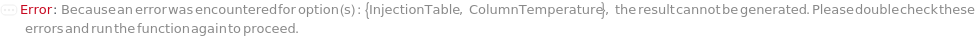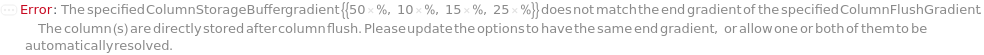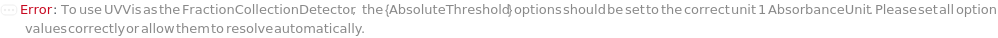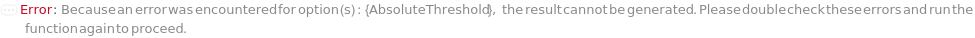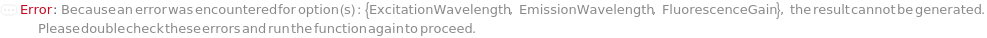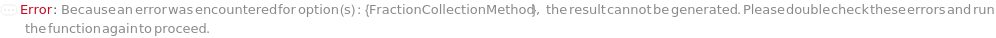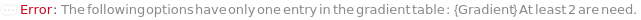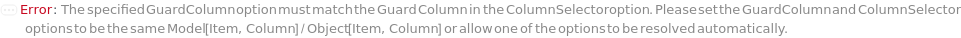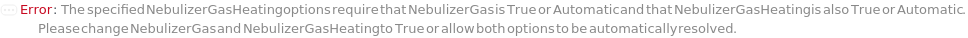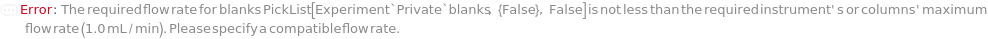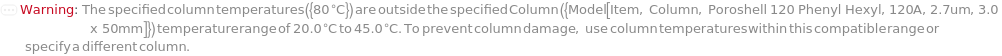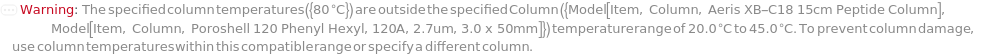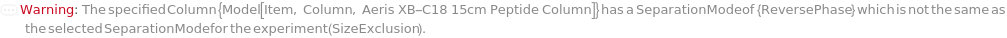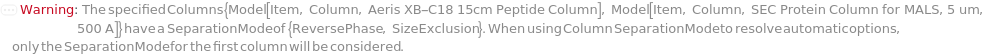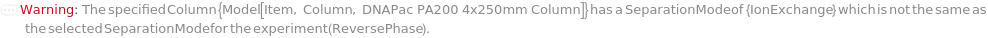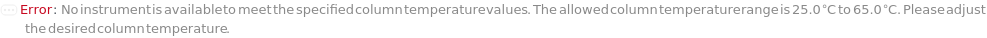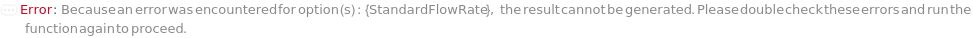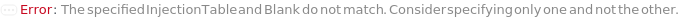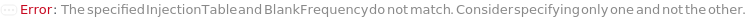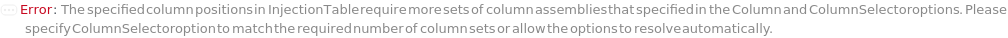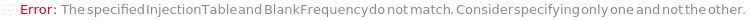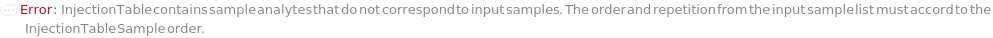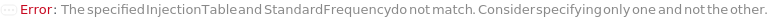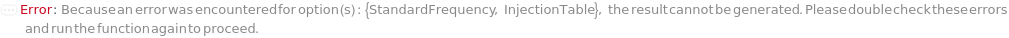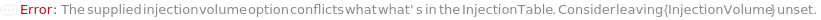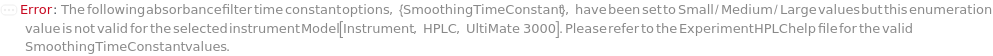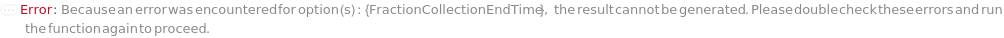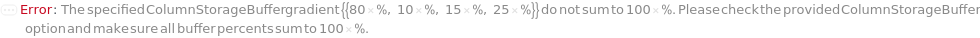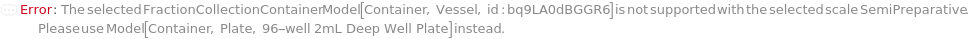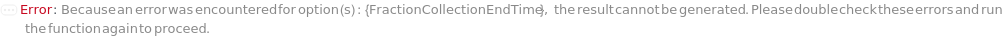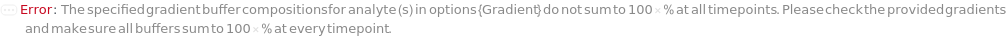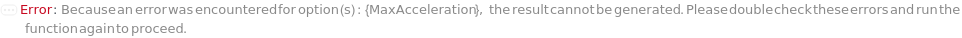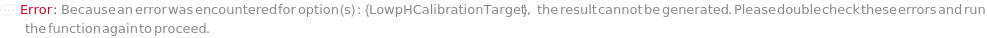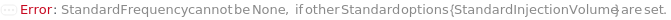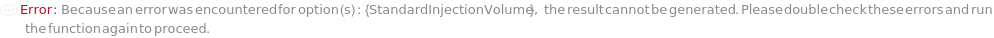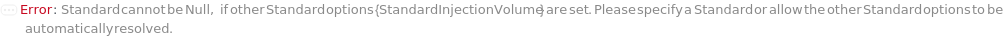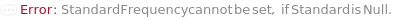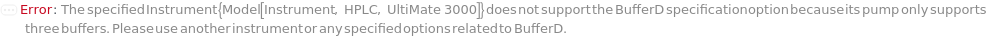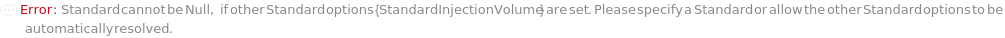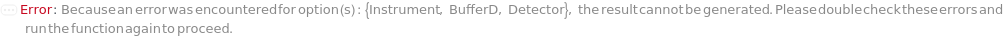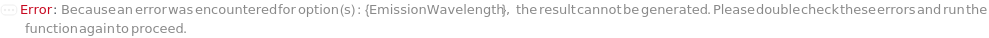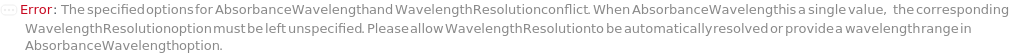General
Instrument
A list of one or more measurement and collection devices to run the experiment on and that satisfy the Scale and Detector options.
Default Calculation: For FractionCollection option specification and/or for Scale->Preparative or SemiPreparative, automatically set to instrument models that have fraction collection capabilities. Otherwise, automatically set to instrument models that meet the requested Detector.
Pattern Description: An object of type or subtype Model[Instrument, HPLC] or Object[Instrument, HPLC] or list of one or more an object of type or subtype Model[Instrument, HPLC] entries or Null.
Programmatic Pattern: ((ObjectP[{Model[Instrument, HPLC], Object[Instrument, HPLC]}] | {ObjectP[Model[Instrument, HPLC]]..}) | Automatic) | Null
Scale
The output of the experiment. Preparative and SemiPreparative indicates that effluent is to be collected by fractions. Analytical indicates that specific measurements will be employed and new SamplesOut will not be generated (e.g the absorbance of the flow with injected sample for a given wavelength).
Default Calculation: If any fraction collection options are specified and injection volume is greater that 500 uL, then Scale -> Preparative; if fraction collection options are specified and injection volume is less than or equal to 500uL, then Scale -> SemiPreparative; otherwise, Scale -> Analytical.
Pattern Description: Preparative, Analytical, or SemiPreparative.
Programmatic Pattern: PurificationScaleP | Automatic
SeparationMode
The category of method used to elicit differential column retention due to interaction between molecules in the mobile phase with those on the stationary phase (column).
Default Calculation: Automatically set to match the Separation Mode listed with the provided column.
Pattern Description: NormalPhase, ReversePhase, IonExchange, SizeExclusion, Affinity, or Chiral.
Programmatic Pattern: SeparationModeP | Automatic
Detector
The type of measurement to employ. Options include Pressure (measures the pump pressure) , Temperature (measures the temperature of the column oven), UVVis (measures the absorbance of a single wavelength of light), PhotoDiodeArray (measures the absorbance of a range of wavelengths), Fluorescence (measures the emitted light from samples after light excitation),pH, Conductance, MultiAngleLightScattering (measures the scattered light intensity at different angles), DynamicLightScattering (measures the scattered light fluctuation), RefractiveIndex (measures how fast light travels through the sample) and EvaporativeLightScattering (separates the flow into airborne droplets and measures the light scattering).
Default Calculation: Automatically set to the detector(s) available for the first selected instrument. For example, if Agilent 1290 Infinity II Instrument is requested, the Detector option will include Pressure and PhotoDiodeArray.
Pattern Description: A selection of one or more of Pressure, Temperature, Conductance, Fluorescence, EvaporativeLightScattering, UVVis, PhotoDiodeArray, CircularDichroism, RefractiveIndex, pH, MultiAngleLightScattering, or DynamicLightScattering.
Programmatic Pattern: DuplicateFreeListableP[Pressure | Temperature | Conductance | Fluorescence | EvaporativeLightScattering | UVVis | PhotoDiodeArray | CircularDichroism | RefractiveIndex | pH | MultiAngleLightScattering | DynamicLightScattering] | Automatic
ColumnSelection
Indicates if multiple different columns will be employed for different samples during the run. All columns are installed during the beginning of the run and the valve on the instrument allows to switch between them automatically.
Default Calculation: If ColumnSelector and InjectionTable are not specified, automatically set to False. If InjectionTable is set but with only one set of column(s), automatically set to False. Otherwise, set to True.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
ColumnPosition
The position of the column selector valve and the desired column configuration that will be used for each sample as it is injected.
Default Calculation: If InjectionTable is specified, automatically set from the Column Position entry for the sample. Otherwise set to PositionA.
Pattern Description: PositionA, PositionB, PositionC, PositionD, PositionE, PositionF, PositionG, or PositionH.
Programmatic Pattern: ColumnPositionP | Automatic
Index Matches to: experiment samples
GuardColumn
The protective device placed in the flow path before the Column in order to adsorb fouling contaminants and, thus, preserve the Column lifetime.
Default Calculation: Automatically set from the column model's PreferredGuardColumn. If Column is Null, GuardColumn is automatically set to Null.
Pattern Description: An object of type or subtype Model[Item, Column], Object[Item, Column], Model[Item, Cartridge, Column], or Object[Item, Cartridge, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column], Model[Item, Cartridge, Column], Object[Item, Cartridge, Column]}] | Automatic) | Null
GuardColumnOrientation
The position of the GuardColumn with respect to the Column, SecondaryColumn and TertiaryColumn. Forward indicates that the GuardColumn will be placed in front of the Column, SecondaryColumn and TertiaryColumn. If a Column is specified and GuardColumnOrientation is Reverse, the GuardColumn will be placed after the Column, SecondaryColumn, and/or TertiaryColumn in the flow path which is typically performed during column cleaning.
Default Calculation: If GuardColumn is specified automatically set to Forward.
Pattern Description: Forward or Reverse or Null.
Programmatic Pattern: (ColumnOrientationP | Automatic) | Null
Column
The item containing the stationary phase through which the mobile phase and input samples flow. It adsorbs and separates the molecules within the sample based on the properties of the mobile phase, Samples, Column material, and the desired column temperature in the specified InjectionTable.
Default Calculation: Automatically set to a column model compatible for the instrument selected and specified separation Mode.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic) | Null
ColumnOrientation
The direction of the Column with respect to the flow. Forward indicates that the Column will be placed in the direction indicated by the column manufacturer for standard operation. Reverse indicates that the Column will be placed in the opposite direction indicated by the column manufacturer for standard operation. This also specifies the orientation of secondary and tertiary columns if provided.
Default Calculation: Automatically set to Forward if column orientation options are not specified.
Pattern Description: Forward or Reverse or Null.
Programmatic Pattern: (ColumnOrientationP | Automatic) | Null
SecondaryColumn
The additional stationary phase through which the mobile phase and input samples flow. The SecondaryColumn selectively adsorb analytes and is connected to flow path, downstream of the Column.
Default Calculation: If ColumnSelector is specified, set from there; otherwise, set to Null.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic) | Null
TertiaryColumn
The additional stationary phase through which the mobile phase and input samples flow. The TertiaryColumn selectively adsorb analytes and is connected to flow path, downstream of the SecondaryColumn.
Default Calculation: If ColumnSelector is specified, set from there; otherwise, set to Null.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic) | Null
IncubateColumn
Indicates if the columns are placed inside the column oven compartment of the HPLC instrument during the experiment. If set to False, the columns are placed on a rack outside the column oven under ambient temperature.
Default Calculation: Automatically set to False if the selected connection of GuardColumn, Column, SecondaryColumn, and TertiaryColumn cannot fit into the column oven compartment of the Instrument. Otherwise set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BufferA
A solvent or buffer placed in the 'A' bottle as shown in Figure 2.1.1 - 2.9.1 of ExperimentHPLC help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientA option.
Default Calculation: Automatically set from the SeparationMode (e.g. Water buffer if ReversePhase) option or the objects specified in the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
BufferB
A solvent or buffer placed in the 'B' bottle as shown in Figure 2.1.1 - 2.9.1 of ExperimentHPLC help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientB option.
Default Calculation: Automatically set from SeparationMode (e.g. Acetonitrile buffer if ReversePhase) or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
BufferC
A solvent or buffer placed in the 'C' bottle as shown in Figure 2.1.1 - 2.9.1 of ExperimentHPLC help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientC option.
Default Calculation: Automatically set from the SeparationMode option or the objects specified in the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
BufferD
A solvent or buffer placed in the 'D' bottle as shown in Figure 2.1.1 - 2.5.1 of ExperimentHPLC help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientD option.
Default Calculation: If the specified Instrument's pump does not support Buffer D, automatically set to Null. Otherwise, set from the SeparationMode option or the objects specified in the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
Column Installation
ColumnSelector
The set of all the columns loaded into the Instrument's column selector and referenced in Column, SecondaryColumn, TertiaryColumn. The Serial configuration indicates one fluid line for all the samples, Standard and Blank. The Selector configuration indicates use of a column selector where the column line is programmatically switchable between samples.
Default Calculation: If ColumnSelection is False, set to match the values in Column, SecondaryColumn, TertiaryColumn, ColumnOrientation, GuardColumn and GuardColumnOrientation options.
Pattern Description: {Column Position, Guard Column, Guard Column Orientation, Column, Column Orientation, Secondary Column, Tertiary Column}
Programmatic Pattern: {ColumnPositionP | Automatic | Null, ObjectP[{Model[Item, Column], Object[Item, Column], Model[Item, Cartridge, Column], Object[Item, Cartridge, Column]}] | (Automatic | Null), ColumnOrientationP | (Automatic | Null), ObjectP[{Model[Item, Column], Object[Item, Column]}] | (Automatic | Null), ColumnOrientationP | (Automatic | Null), ObjectP[{Model[Item, Column], Object[Item, Column]}] | (Automatic | Null), ObjectP[{Model[Item, Column], Object[Item, Column]}] | (Automatic | Null)} | Automatic
Index Matches to: ColumnSelector
Separation
ColumnStorageBuffer
The solvent or gradient at the end of the column flush in which the column will be stored in long term after removed from the instrument. The provided solvent option must match one of the buffers used in the experiment and the column flush will end with 100% gradient of the selected buffer. If a gradient with percents of buffers is specified, the column flush will end with the specified gradient composition.
Default Calculation: Automatically set to the last gradient composition of the ColumnFlushGradient option if provided. Otherwise set from the StorageBuffer field from the Model[Item,Column] specified for the specified Column, if it is one of the buffers specified in the protocol.
Pattern Description: Buffer or Gradient or Null.
Programmatic Pattern: (((ObjectP[{Object[Sample], Model[Sample]}] | _String) | {RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent]}) | Automatic) | Null
Index Matches to: ColumnSelector
GradientA
The composition of BufferA within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientB, GradientC, and GradientD options such that the total amounts to 100%.
Pattern Description: Binary or Isocratic.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic
Index Matches to: experiment samples
GradientB
The composition of BufferB within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientA, GradientC, and GradientD options such that the total amounts to 100%.
Pattern Description: Binary or Isocratic.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic
Index Matches to: experiment samples
GradientC
The composition of BufferC within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientA, GradientB, and GradientD options such that the total amounts to 100%.
Pattern Description: Binary or Isocratic.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic
Index Matches to: experiment samples
GradientD
The composition of BufferD within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientA, GradientB, and GradientC options such that the total amounts to 100%.
Pattern Description: Binary or Isocratic or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: experiment samples
FlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the gradient options. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If Gradient option is specified, automatically set from the method given in the Gradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 200 milliliters per minute or list of one or more {Time, Flow Rate} entries.
Programmatic Pattern: (RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)]}..}) | Automatic
Index Matches to: experiment samples
MaxAcceleration
When ramping up the FlowRate of solvent through the instrument, the maximum allowed change per time in the FlowRate.
Default Calculation: For Waters and Agilent instruments, automatically set to the lowest value from Max the Column, Instrument, and GuardColumn models. For other instruments, automatically set to Null.
Pattern Description: Greater than 0 milliliters per minute squared or Null.
Programmatic Pattern: (GreaterP[0*(Milliliter/Minute/Minute)] | Automatic) | Null
Gradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow. Specific parameters of a gradient object can be overridden by specific options. Differential Refractive Index Reference Loading refers to the HPLC refractive index loading reference values as shown in the Figure 2.7.4. When open, the flow downstream of the column is loaded into the two flow cells simultaneously.
Default Calculation: Automatically set to best meet all the Gradient options (e.g. GradientA, GradientB, GradientC, GradientD, FlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate, Differential Refractive Index Reference Loading} entries.
Programmatic Pattern: (ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)], Open | Closed | None | Automatic}..}) | Automatic
Index Matches to: experiment samples
Sample Parameters
NumberOfReplicates
The number of times to repeat measurements on each provided sample(s). If Aliquot -> True, this also indicates the number of times each provided sample will be aliquoted. For experiment samples {A,B,C} if NumberOfReplicates is specified as 3, the order of samples to run on the instrument will be {A,A,A,B,B,B,C,C,C}.
Pattern Description: Greater than or equal to 1 and less than or equal to 96 in increments of 1 or Null.
Programmatic Pattern: RangeP[1, 96, 1] | Null
InjectionTable
The order of Sample, Standard and Blank sample injected into the Instrument during measurement and/or collection. This also includes the priming and flushing of the column(s).
Default Calculation: Samples are inserted in the order of the input samples based with the number of replicates. Standard and Blank samples are inserted based on the determination of StandardFrequency and BlankFrequency options. For example, StandardFrequency -> FirstAndLast and BlankFrequency -> Null result in Standard samples injected first, then samples, and then the Standard sample set again at the end. Column priming is inserted at the beginning and repeated according to ColumnPrimeFrequency. Column flushing is inserted at the end.
Pattern Description: List of one or more {Type, Sample, InjectionVolume, Column Position, Column Temperature, Gradient} entries.
Programmatic Pattern: {{Standard | Sample | Blank | ColumnPrime | ColumnFlush, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | (Automatic | Null), RangeP[0*Microliter, 16*Milliliter] | (Automatic | Null), ColumnPositionP | Automatic, RangeP[5*Celsius, 90*Celsius] | (Ambient | Automatic), ObjectP[Object[Method, Gradient]] | Automatic}..} | Automatic
SampleTemperature
The temperature of the chamber in which the samples, Standard, and Blank are stored while waiting for the Injection.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 40 degrees Celsius.
Programmatic Pattern: RangeP[5*Celsius, 40*Celsius] | Ambient
ColumnTemperature
The temperature of the column assembly throughout the measurement and/or fraction collection.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the Gradient option or the column temperature for the sample in the InjectionTable option; otherwise, set to Ambient (no column oven temperature control).
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 90 degrees Celsius.
Programmatic Pattern: (RangeP[5*Celsius, 90*Celsius] | Ambient) | Automatic
Index Matches to: experiment samples
InjectionVolume
The physical quantity of sample loaded into the flow path for measurement and/or collection.
Default Calculation: Automatically defaults to the lesser of 10 uL or 90% of the available sample volume for Analytical measurement, lesser of 500 uL or 90% of the available sample volume for Semipreparative measurement and lesser of 5mL or 90% of available sample volume for Preparative measurement.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 16 milliliters.
Programmatic Pattern: RangeP[0*Microliter, 16*Milliliter] | Automatic
Index Matches to: experiment samples
NeedleWashSolution
The solvent used to wash the injection needle before each sample introduction. For Dionex instruments, this is the same as BufferC and cannot be defined separately.
Default Calculation: If the instrument shares NeedleWashSolution with BufferC, automatically set to specified BufferC. Otherwise, defaults to Model[Sample, "Milli-Q water"] for IonExchange and SizeExclusion SeparationType or Model[Sample, StockSolution, "20% Methanol in MilliQ Water"] for other SeparationType.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample]
Programmatic Pattern: ObjectP[{Object[Sample], Model[Sample]}] | Automatic
Detection
AbsorbanceWavelength
The wavelength of light passed through the flow cell for the UVVis Detector. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: If a UVVis Detector is selected or available on the selected instrument, automatically set to the absorbance wavelength corresponding to the maximum extinction coefficient from the ExtinctionCoefficients field in the identity model of the samples specified. If no ExtinctionCoefficients available, automatically set to to 260 Nanometer for oligomers or 280 Nanometer for proteins. If a PhotoDiodeArray Detector is selected or available on the selected Instrument, automatically set to All.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 950*Nanometer] | All | RangeP[190*Nanometer, 950*Nanometer] ;; RangeP[200*Nanometer, 950*Nanometer]) | Automatic) | Null
Index Matches to: experiment samples
WavelengthResolution
The increment in wavelength for the range of wavelength of light passed through the flow for absorbance measurement of PhotoDiodeArray Detector.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected Instrument, automatically set to 2.4 Nanometer.
Pattern Description: Greater than or equal to 0.1 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[0.1*Nanometer, 12.*Nanometer] | Automatic) | Null
Index Matches to: experiment samples
UVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the sample for the PhotoDiodeArray Detector.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected Instrument, automatically set to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
AbsorbanceSamplingRate
The number of times an absorbance measurement is made per second by the detector on the selected instrument. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set to 20/Second.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 120 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 120*(1/Second)] | Automatic) | Null
Index Matches to: experiment samples
SmoothingTimeConstant
The time window used on the instrument software for data filtering in absorbance collection, affecting baseline smoothing and peak height degradation. Raw absorbance signals within the time window are smoothed using a weighted moving average, with the result applied to the leftmost point of the window to effectively suppress high-frequency noise. Optimizing this parameter improves signal-to-noise ratio: shorter time constants give faster response and sharper peaks, while longer ones enhance sensitivity by reducing noise. For Waters instruments, the setting can be a numeric value or an enumeration - Small, Medium, or Large - corresponding to 1, 2, or 4 divided by AbsorbanceSamplingRate. Dionex instruments require a numeric value between 0.01 and 4.55 Second. Agilent instruments accept only specific values based on AbsorbanceSamplingRate. For details, please refer to Figure 3.1 in the ExperimentHPLC help file, and the SmoothingTimeConstants field in Model[Instrument, HPLC].
Pattern Description: Greater than or equal to 0.0125 seconds and less than or equal to 5 seconds or Small, Medium, or Large or Null.
Programmatic Pattern: (HPLCSmoothingTimeConstantP | RangeP[0.0125*Second, 5*Second]) | Null
Index Matches to: experiment samples
ExcitationWavelength
The wavelength(s) of light that is used to excite fluorescence in the samples when passed through the Fluorescence Detector.
Default Calculation: If Fluorescence Detector is selected, automatically set from the FluorescenceExcitationMaximums field in the identity Model of the sample specified.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: experiment samples
EmissionWavelength
The wavelength(s) of light at which fluorescence emitted from the sample is measured in the Fluorescence Detector.
Default Calculation: If Fluorescence Detector is selected, automatically set from the FluorescenceEmissionMaximums field in the identity Model of the sample specified.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: experiment samples
EmissionCutOffFilter
The cut-off wavelength to pre-select the emitted light from the sample and allow only the light with wavelength above the desired value to pass, before the light enters emission monochromator for final wavelength selection for Ultimate 3000 with FLR Detector. If set to None, no cut-off filter is used.
Default Calculation: If a Fluorescence Detector with a cut-off filter wheel is selected, automatically set to None.
Pattern Description: 280 nanometers, 370 nanometers, 435 nanometers, 530 nanometers, or None or Null.
Programmatic Pattern: (HPLCEmissionCutOffFilterP | Automatic) | Null
Index Matches to: experiment samples
FluorescenceGain
For each ExcitationWavelength/EmissionWavelength pair, the signal amplification factor which modulates the percentage of maximum voltage that can be applied to the Photomultiplier Tube of the Fluorescence Detector. Linear increase in voltage applied to the Photomultiplier tube leads to an exponential change in RFU signal. Variable Fluorescence Sensitivity implies a different fluorescence sensitivity for each Excitation/Emission Wavelength pair.
Default Calculation: If the "Ultimate 3000 with FLR Detector" or "Waters Acquity UPLC H-Class FLR" instrument is selected, automatically set to 100 Percent. If the "Agilent 1260 Infinity II Semi-Preparative HPLC with UV/Vis Diode Array and Fluorescence Detectors" instrument is selected, automatically set to 60 Percent.
Pattern Description: Constant or Variable Fluorescence Sensitivity or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {RangeP[0*Percent, 100*Percent]..}) | Automatic) | Null
Index Matches to: experiment samples
FluorescenceFlowCellTemperature
The temperature that the thermostat inside the fluorescence flow cell of the Fluorescence Detector is set to during the fluorescence measurement of the sample.
Default Calculation: If Fluorescence Detector is selected and temperature control is available on that unit, automatically set to Ambient.
Pattern Description: Ambient or greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[25*Celsius, 50*Celsius] | Ambient) | Automatic) | Null
Index Matches to: experiment samples
LightScatteringLaserPower
The laser power filter used in the Multi-Angle static Light Scattering (MALS) and Dynamic Light Scattering (DLS) Detector. 100% means that no filter is being used to restrict the laser power.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to 100 Percent.
Pattern Description: Greater than or equal to 10 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[10*Percent, 100*Percent] | Automatic) | Null
Index Matches to: experiment samples
LightScatteringFlowCellTemperature
The temperature that the thermostat inside the flow cell of the Detector is set to during the Multi-Angle static Light Scattering (MALS) and/or Dynamic Light Scattering (DLS) measurement of the sample.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to Ambient.
Pattern Description: Ambient or greater than or equal to 20 degrees Celsius and less than or equal to 70 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[20*Celsius, 70*Celsius] | Ambient) | Automatic) | Null
Index Matches to: experiment samples
RefractiveIndexMethod
The type of refractive index measurement of the Refractive Index (RI) Detector for the measurement of the sample. When DifferentialRefractiveIndex is selected, the refractive index difference between the flow downstream sample and the reference solvent is measured. See Figure 2.7.4 for more information.
Default Calculation: If RefractiveIndex Detector is selected and Differential Refractive Index Reference Loading is set to Closed in Gradient, automatically set to DifferentialRefractiveIndex. Otherwise automatically set to RefractiveIndex.
Pattern Description: RefractiveIndex or DifferentialRefractiveIndex or Null.
Programmatic Pattern: ((RefractiveIndex | DifferentialRefractiveIndex) | Automatic) | Null
Index Matches to: experiment samples
RefractiveIndexFlowCellTemperature
The temperature that the thermostat inside the refractive index flow cell of the Refractive Index (RI) Detector is set to during the refractive index measurement of the sample.
Default Calculation: If RefractiveIndex Detector is selected, automatically set to Ambient.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 65 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[4*Celsius, 65*Celsius] | Ambient) | Automatic) | Null
Index Matches to: experiment samples
pHCalibration
Indicates if 2-point calibration of the pH probe should be performed before the experiment starts. pH And Conductivity calibration is performed monthly every time a qualification procedure is run on the instrument.
Default Calculation: Automatically set to True if pH Detector is selected.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
LowpHCalibrationBuffer
The low pH buffer that should be used to calibrate the pH probe in the 2-point calibration.
Default Calculation: Automatically set to Model[Sample, "pH 4.01 Calibration Buffer, Sachets"] if pH Detector is selected and pHCalibration is True.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
LowpHCalibrationTarget
The pH of the LowpHCalibrationBuffer that should be used to calibrate the pH probe in the 2-point calibration.
Default Calculation: Automatically set to the pH of the LowpHCalibrationBuffer's model.
Pattern Description: Greater than or equal to 0 and less than or equal to 14 or Null.
Programmatic Pattern: (RangeP[0, 14] | Automatic) | Null
HighpHCalibrationBuffer
The high pH buffer that should be used to calibrate the pH probe in the 2-point calibration.
Default Calculation: Automatically set to Model[Sample, "pH 7.00 Calibration Buffer, Sachets"] if pH Detector is selected and pHCalibration is True. If HighpHCalibrationTarget is specified, set to a buffer with pH value close to that.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
HighpHCalibrationTarget
The pH of the HighpHCalibrationBuffer that should be used to calibrate the pH probe in the 2-point calibration.
Default Calculation: Automatically set to the pH of the HighpHCalibrationBuffer's model.
Pattern Description: Greater than or equal to 0 and less than or equal to 14 or Null.
Programmatic Pattern: (RangeP[0, 14] | Automatic) | Null
pHTemperatureCompensation
Indicates if the measured pH value should be automatically corrected according to the temperature inside the pH flow cell.
Default Calculation: Automatically set to True if pH Detector is selected.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
ConductivityCalibration
Indicates if 1-point calibration of the conductivity probe should be performed before the experiment starts. pH And Conductivity calibration is performed monthly every time a qualification procedure is run on the instrument.
Default Calculation: Automatically set to True if Conductivity Detector is selected.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
ConductivityCalibrationBuffer
The buffer that should be used to calibrate the conductivity probe in the 1-point calibration.
Default Calculation: Automatically set to Model[Sample, "Conductivity Standard 1413
µS, Sachets"] if Conductivity Detector is selected and ConductivityCalibration is True.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
ConductivityCalibrationTarget
The conductivity value of the ConductivityCalibrationBuffer that should be used to calibrate the conductivity probe in the 1-point calibration.
Default Calculation: Automatically set to the Conductivity of the ConductivityCalibrationBuffer's model.
Pattern Description: Greater than or equal to 10 microsiemens per centimeter and less than or equal to 1000 millisiemens per centimeter or Null.
Programmatic Pattern: (RangeP[10*Micro*(Siemens/Centimeter), 1000*Milli*(Siemens/Centimeter)] | Automatic) | Null
ConductivityTemperatureCompensation
Indicates if the measured conductivity value should be automatically corrected according to the temperature inside the conductivity flow cell.
Default Calculation: Automatically set to True if Conductivity Detector is selected.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
NebulizerGas
Indicates if Nitrogen sheath gas is flowed along with the sample within the EvaporativeLightScattering Detector.
Default Calculation: If EvaporativeLightScattering detector is selected, automatically set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
NebulizerGasHeating
Indicates if the sheath gas that carries samples in the EvaporativeLightScattering Detector is heated.
Default Calculation: If EvaporativeLightScattering Detector is selected and NebulizerGas is True, automatically set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
NebulizerHeatingPower
The relative magnitude of the heating element used to heat the sheath gas for the EvaporativeLightScattering Detector (Corresponding temperature not measured by the manufacturer). Higher percent values correspond to percent of power going to heating coil.
Default Calculation: If EvaporativeLightScattering Detector is selected and NebulizerGasHeating is True, automatically set to 50 Percent.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: experiment samples
NebulizerGasPressure
The applied pressure of sheath gas for the EvaporativeLightScattering Detector (Flow rate unmeasured by the manufacturer). Higher pressure (20-60 PSI) corresponds to faster sheath gas flow.
Default Calculation: If EvaporativeLightScattering Detector is selected and NebulizerGas is True, automatically set to 40 PSI.
Pattern Description: Greater than or equal to 20 pounds
‐force per inch squared and less than or equal to 60 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[20*PSI, 60*PSI] | Automatic) | Null
Index Matches to: experiment samples
DriftTubeTemperature
The set temperature of the chamber thermostat through which the nebulized analytes flow within the EvaporativeLightScattering Detector. The purpose to heat the drift tube is to evaporate any unevaporated solvent remaining in the flow from the nebulizer.
Default Calculation: If EvaporativeLightScattering Detector is selected and NebulizerGas is True, automatically set to 50 Celsius.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 100*Celsius] | Automatic) | Null
Index Matches to: experiment samples
ELSDGain
The percent of maximum voltage sent to the Photo Multiplier Tube (PMT) for signal amplification for the EvaporativeLightScattering measurement. The percentage value specified here is converted into a unitless factor from 0 to 1000 which the software accepts to modulate the voltage for the PMT.
Default Calculation: If EvaporativeLightScattering Detector is selected and NebulizerGas is True, automatically set to 50 Percent.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: experiment samples
ELSDSamplingRate
The frequency of evaporative light scattering measurement. Lower values will be less susceptible to noise but will record less frequently across time. Lower or higher values do not affect the y axis of the measurement.
Default Calculation: If EvaporativeLightScattering Detector is selected and NebulizerGas is True, automatically set to 20/Second.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second)] | Automatic) | Null
Index Matches to: experiment samples
Fraction Collection
CollectFractions
Indicates if effluents from the Column should be captured and stored at specific time windows or during detection of peaks (fractions).
Default Calculation: If Scale is Preparative/SemiPreparative or any fraction collection options are specified, set to True. For analytical measurements, set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
SamplesOutUltrasonicIncompatible
Indicates if the samples collected during the elution of this sample should get UltrasonicIncompatible property set. Null indicates that the UltrasonicIncompatible will be resolved from the composition of the fraction. If CollectFractions is set to False, this option is ignored.
Pattern Description: True or False or Null.
Programmatic Pattern: BooleanP | Null
FractionCollectionDetector
The type of measurement that is used as signal to trigger fraction collection. It corresponds to the type of detector on the instrument.
Default Calculation: If CollectFractions is True, automatically set to the Detector in the Detector option for which the Detector hardware can communicate with the fraction collection (as indicated in the Instrumentation Table of ExperimentHPLC help file).
Pattern Description: Fluorescence, UVVis, PhotoDiodeArray, pH, or Conductance or Null.
Programmatic Pattern: (HPLCFractionCollectionDetectorTypeP | Automatic) | Null
FractionCollectionContainer
The container in which the fractions are collected on the selected instrument's fraction collector.
Default Calculation: Automatically set to Model[Container, Plate, "96-well 2mL Deep Well Plate"] for UltiMate 3000 HPLC instruments and to Model[Container, Vessel, "50mL Tube"] for Agilent 1290 Infinity II instrument.
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[{Model[Container]}] | Automatic) | Null
FractionCollectionMethod
The fraction collection method object which describes the conditions for which a fraction is collected. Specific parameters of the object can be overridden by other fraction collection options.
Pattern Description: An object of type or subtype Object[Method, FractionCollection] or Null.
Programmatic Pattern: ObjectP[Object[Method, FractionCollection]] | Null
Index Matches to: experiment samples
FractionCollectionStartTime
The time at which to start column effluent capture. Time in this case is the duration from the start of sample injection.
Default Calculation: Automatically set from the method specified in the FractionCollectionMethod option, if available. Otherwise set to the second time point of the gradient domains if there are more than two time points, or the first time point if not.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
FractionCollectionEndTime
The time to end column effluent capture. Time in this case is the duration from the start of sample injection.
Default Calculation: Automatically inherited from the method specified in the FractionCollectionMethod option. Otherwise set to the last time point of the gradient domains.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
FractionCollectionMode
The method by which fractions collection should be triggered (peak detection, a constant threshold, or a fixed fraction time). In Peak detection mode, the fraction collection is triggered when a change in slope of the FractionCollectionDetector signal is observed for a specified PeakSlopeDuration time. In constant Threshold mode, whenever the signal from the FractionCollectionDetector is above the specified value, fraction collection is triggered. In fixed fraction Time mode, fractions are collected during the whole time interval specified.
Default Calculation: Automatically inherited from a method specified by FractionCollectionMethod option, or implicitly resolved from other fraction collection options. If AbsoluteThreshold is specified, set to Threshold. If PeakSlope is specified, set to Peak. If MaxCollectionPeriod is specified, set to Time. Otherwise set to Threshold if CollectFractions is True.
Pattern Description: Time, Peak, or Threshold or Null.
Programmatic Pattern: (FractionCollectionModeP | Automatic) | Null
Index Matches to: experiment samples
MaxFractionVolume
The maximum amount of sample to be collected in a single fraction. If fraction detection trigger is not off, the collector moves position to the next container. For example, if AbsorbanceThreshold is set to 180 MilliAbsorbanceUnit and at MaxFractionVolume the absorbance value is still above 180 MilliAbsorbanceUnit, the fraction collector continues to collect fractions in the next container in line.
Default Calculation: If FractionCollection is True, automatically set according to the MaxFractionVolume in the method specified by FractionCollectionMethod option, if available. If FractionCollectionContainer is specified, set to MaxVolume of the Model specified. Otherwise, automatically set to 1.8 Milliliter for UltiMate 3000 HPLC instruments and 45 Milliliter for Agilent 1290 Infinity II instrument.
Pattern Description: Greater than or equal to 10 microliters and less than or equal to 50 milliliters or Null.
Programmatic Pattern: (RangeP[10*Microliter, 50*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
MaxCollectionPeriod
The amount of time after which a new fraction will be generated (Fraction Collector moves to the next vial) when FractionCollectionMode is Time. For example, if MaxCollectionPeriod is 120 Second, the fraction collector continues to collect fractions in the next container in line after 120 Second.
Default Calculation: If FractionCollection is True, automatically set according to the MaxCollectionPeriod in the method specified by FractionCollectionMethod option, if available. Otherwise automatically set to the time it takes to fill to the MaxFractionVolume based on the flow rates.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Second, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
AbsoluteThreshold
The signal value from FractionCollectionDetector above which fractions will always be collected, when FractionCollectionMode is Threshold.
Default Calculation: Inherited from a method specified by FractionCollectionMethod option or set based on FractionCollectionDetector if FractionCollectionMode is Threshold. If the FractionCollectionDetector is UVVis, automatically set to 500 Milli AbsorbanceUnit. If the FractionCollectionDetector is Fluorescence, automatically set to 100 Milli RFU.
Pattern Description: Others or pH or Null.
Programmatic Pattern: ((RangeP[0, 14] | (GreaterEqualP[0*AbsorbanceUnit] | GreaterEqualP[0*RFU] | GreaterEqualP[10*Micro*(Siemens/Centimeter)])) | Automatic) | Null
Index Matches to: experiment samples
PeakSlope
The minimum slope (signal change per second) required for PeakSlopeDuration to trigger peak detection and start fraction collection. Fraction collection end slope is defined as the opposite of PeakSlope and fraction collection will continue until the slope exceeds the negative of the PeakSlope. For instance, if PeakSlope is set to 1 Milli Absorbance Unit/Second, fraction collection begins when the slope surpasses this value and ends when the slope falls below -1 Milli Absorbance Unit/Second. If a PeakEndThreshold is specified, both the PeakEndThreshold and PeakSlope conditions must be satisfied to stop fraction collection. A new peak and corresponding fraction can be registered when the slope exceeds the PeakSlope again.
Default Calculation: If FractionCollection is True, automatically set according to the PeakSlope in the method specified by FractionCollectionMethod option, if available. If the FractionCollectionDetector is UVVis and FractionCollectionMode is Peak, automatically set to 1 Milli AbsorbanceUnit/Second. If the FractionCollectionDetector is Fluorescence and FractionCollectionMode is Peak, automatically set to 0.2 Milli RFU/Second.
Pattern Description: Others or pH or Null.
Programmatic Pattern: ((GreaterEqualP[0*(1/Second)] | (GreaterEqualP[0*(AbsorbanceUnit/Second)] | GreaterEqualP[0*(RFU/Second)] | GreaterEqualP[10*Micro*(Siemens/Centimeter/Second)])) | Automatic) | Null
Index Matches to: experiment samples
PeakSlopeDuration
The minimum duration that changes in slopes must be maintained before fraction collection is registered or ended. This option is only applicable for UltiMate 3000 HPLC instruments.
Default Calculation: If FractionCollection is True, automatically set according to the PeakSlopeDuration in the method specified by FractionCollectionMethod option, if available. Otherwise automatically set to 0.5 Second if FractionCollectionMode is Peak.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 4 seconds or Null.
Programmatic Pattern: (RangeP[0*Second, 4*Second] | Automatic) | Null
Index Matches to: experiment samples
PeakEndThreshold
The signal value below which the end of a peak is marked and fraction collection stops when FractionCollectionMode is Peak. Both the PeakEndThreshold and PeakSlope conditions must be satisfied to stop fraction collection. This option is only applicable for UltiMate 3000 HPLC instruments.
Default Calculation: If FractionCollection is True, automatically set according to the PeakEndThreshold in the method specified by FractionCollectionMethod option, if available. If FractionCollectionMode is Peak, automatically set to 1 Milli AbsorbanceUnit for UVVis detector, 0.2 Milli * RFU for Fluorescence detector, 10 for pH detector or 10.0 Milli * Siemens / Centimeter for Conductivity detector.
Pattern Description: Others or pH or Null.
Programmatic Pattern: ((RangeP[0, 14] | (GreaterEqualP[0*AbsorbanceUnit] | GreaterEqualP[0*RFU] | GreaterEqualP[10*Micro*(Siemens/Centimeter)])) | Automatic) | Null
Index Matches to: experiment samples
Standards
Standard
The reference compound(s) to inject to the instrument, often used for quantification or to check internal measurement consistency.
Default Calculation: If any other Standard option is specified, automatically set based on the SeparationMode option. If InjectionTable is specified, set from the specified Standard entries in the InjectionTable.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardInjectionVolume
The physical quantity of Standard sample loaded into the flow path on the selected instrument along with the mobile phase onto the stationary phase.
Default Calculation: Automatically set equal to the first entry in InjectionVolume.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 16 milliliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 16*Milliliter] | Automatic) | Null
Index Matches to: Standard
StandardFrequency
The frequency at which Standard measurements will be inserted between the experiment samples.
Default Calculation: If injectionTable is given, automatically set to Null and the sequence of Standards specified in the InjectionTable will be used in the experiment. If any other Standard option is specified, automatically set to FirstAndLast.
Pattern Description: Greater than 0 in increments of 1 or None, First, Last, FirstAndLast, or GradientChange or Null.
Programmatic Pattern: (((None | First | Last | FirstAndLast | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
StandardColumnPosition
The position of the column selector valve and the desired column configuration that will be used for each standard sample as it is injected.
Default Calculation: If InjectionTable is specified, automatically set from the Column Position entry for the standard sample. Otherwise set to PositionA.
Pattern Description: PositionA, PositionB, PositionC, PositionD, PositionE, PositionF, PositionG, or PositionH or Null.
Programmatic Pattern: (ColumnPositionP | Automatic) | Null
Index Matches to: Standard
StandardColumnTemperature
The temperature of the column assembly throughout the Standard gradient and measurement.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the StandardGradient option or the column temperature for the sample in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[5*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
Index Matches to: Standard
StandardGradientA
The composition of BufferA within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientB, StandardGradientC, and StandardGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradientB
The composition of BufferB within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientA, StandardGradientC, and StandardGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradientC
The composition of BufferC within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientA, StandardGradientB, and StandardGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradientD
The composition of BufferD within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientA, StandardGradientB, and StandardGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the StandardGradient options during Standard measurement. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If StandardGradient option is specified, automatically set from the method given in the StandardGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 200 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow for Standard measurement. Specific parameters of a gradient object can be overridden by specific options. Differential Refractive Index Reference Loading refers to the HPLC refractive index loading reference values as shown in the Figure 2.7.4. When open, the flow downstream of the column is loaded into the two flow cells simultaneously.
Default Calculation: Automatically set to best meet all the StandardGradient options (e.g. StandardGradientA, StandardGradientB, StandardGradientC, StandardGradientD, StandardFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate, Differential Refractive Index Reference Loading} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)], Open | Closed | None | Automatic}..}) | Automatic) | Null
Index Matches to: Standard
StandardAbsorbanceWavelength
For Standard measurement, the wavelength of light passed through the flow cell for the UVVis Detector. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the same value as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 950*Nanometer] | All | RangeP[190*Nanometer, 950*Nanometer] ;; RangeP[200*Nanometer, 950*Nanometer]) | Automatic) | Null
Index Matches to: Standard
StandardWavelengthResolution
The increment in wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector for Standard measurement.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected Instrument, automatically set equal to the same value as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 0.1 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[0.1*Nanometer, 12.*Nanometer] | Automatic) | Null
Index Matches to: Standard
StandardUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the sample for the PhotoDiodeArray Detector for Standard measurement.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected Instrument, automatically set to the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardAbsorbanceSamplingRate
The number of times an absorbance measurement is made per second for Standard sample. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 0.2 reciprocal seconds and less than or equal to 120 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[0.2*(1/Second), 120*(1/Second)] | Automatic) | Null
Index Matches to: Standard
StandardSmoothingTimeConstant
The time window used on the instrument software for data filtering in absorbance collection of Standard sample, affecting baseline smoothing and peak height degradation. Raw absorbance signals within the time window are smoothed using a weighted moving average, with the result applied to the leftmost point of the window to effectively suppress high-frequency noise. Optimizing this parameter improves signal-to-noise ratio: shorter time constants give faster response and sharper peaks, while longer ones enhance sensitivity by reducing noise. For Waters instruments, the setting can be a numeric value or an enumeration - Small, Medium, or Large - corresponding to 1, 2, or 4 divided by AbsorbanceSamplingRate. Dionex instruments require a numeric value between 0.01 and 4.55 Second. Agilent instruments accept only specific values based on AbsorbanceSamplingRate. For details, please refer to Figure 3.1 in the ExperimentHPLC help file, and the SmoothingTimeConstants field in Model[Instrument, HPLC].
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is requested on a Waters HPLC instrument, automatically set to the first entry in SmoothingTimeConstant.
Pattern Description: Greater than or equal to 0.0125 seconds and less than or equal to 5 seconds or Small, Medium, or Large or Null.
Programmatic Pattern: ((HPLCSmoothingTimeConstantP | RangeP[0.0125*Second, 5*Second]) | Automatic) | Null
Index Matches to: Standard
StandardExcitationWavelength
The wavelength(s) that is used to excite fluorescence in the Standard sample in the Fluorescence Detector.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in ExcitationWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: Standard
StandardEmissionWavelength
The wavelength(s) of light at which fluorescence emitted from the Standard sample is measured in the Fluorescence Detector.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in EmissionWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: Standard
StandardEmissionCutOffFilter
The cut-off wavelength to pre-select the emitted light from the Standard sample and allow only the light with wavelength above the desired value to pass, before the light enters emission monochromator for final wavelength selection for Ultimate 3000 with FLR Detector. If set to None, no cut-off filter is used.
Default Calculation: If a Fluorescence Detector with a cut-off filter wheel is selected, automatically set to the first entry in EmissionCutOffFilter.
Pattern Description: 280 nanometers, 370 nanometers, 435 nanometers, 530 nanometers, or None or Null.
Programmatic Pattern: (HPLCEmissionCutOffFilterP | Automatic) | Null
Index Matches to: Standard
StandardFluorescenceGain
For each StandardExcitationWavelength/StandardEmissionWavelength pair, the signal amplification factor which modulates the percentage of maximum voltage that can be applied to the Photomultiplier Tube of the Fluorescence Detector during Standard measurement. Linear increase in voltage applied to the Photomultiplier tube leads to an exponential change in RFU signal. Variable Fluorescence Sensitivity implies a different fluorescence sensitivity for each Excitation/Emission Wavelength pair.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in FluorescenceGain.
Pattern Description: Constant or Variable Fluorescence Sensitivity or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {RangeP[0*Percent, 100*Percent]..}) | Automatic) | Null
Index Matches to: Standard
StandardFluorescenceFlowCellTemperature
The temperature that the thermostat inside the fluorescence flow cell of the Fluorescence Detector is set to during the fluorescence measurement of the Standard sample.
Default Calculation: If Fluorescence Detector is selected and temperature control is available on that unit, automatically set to the first entry in FluorescenceFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[25*Celsius, 50*Celsius] | Ambient) | Automatic) | Null
Index Matches to: Standard
StandardLightScatteringLaserPower
The laser power filter used in the Multi-Angle static Light Scattering (MALS) and Dynamic Light Scattering (DLS) Detector for the measurement of the Standard sample. 100% means that no filter is being used to restrict the laser power.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to the first entry in LightScatteringLaserPower.
Pattern Description: Greater than or equal to 10 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[10*Percent, 100*Percent] | Automatic) | Null
Index Matches to: Standard
StandardLightScatteringFlowCellTemperature
The temperature that the thermostat inside the flow cell of the Detector is set to during the Multi-Angle static Light Scattering (MALS) and/or Dynamic Light Scattering (DLS) measurement of the Standard sample.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to the first entry in LightScatteringFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to -15 degrees Celsius and less than or equal to 210 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[-15*Celsius, 210*Celsius] | Ambient) | Automatic) | Null
Index Matches to: Standard
StandardRefractiveIndexMethod
The type of refractive index measurement of the Refractive Index (RI) Detector for the measurement of the Standard. When DifferentialRefractiveIndex is selected, the refractive index difference between the flow downstream sample and the reference solvent is measured. See Figure 2.7.4 for more information.
Default Calculation: If RefractiveIndex Detector is selected and Differential Refractive Index Reference Loading is set to Closed in StandardGradient, automatically set to DifferentialRefractiveIndex. Otherwise automatically set to RefractiveIndex.
Pattern Description: RefractiveIndex or DifferentialRefractiveIndex or Null.
Programmatic Pattern: ((RefractiveIndex | DifferentialRefractiveIndex) | Automatic) | Null
Index Matches to: Standard
StandardRefractiveIndexFlowCellTemperature
The temperature that the thermostat inside the refractive index flow cell of the Refractive Index (RI) Detector is set to during the refractive index measurement of the Standard sample.
Default Calculation: If RefractiveIndex Detector is selected, automatically set to the first entry in RefractiveIndexFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 65 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[4*Celsius, 65*Celsius] | Ambient) | Automatic) | Null
Index Matches to: Standard
StandardNebulizerGas
Indicates if Nitrogen sheath gas is flowed along with the Standard sample within the EvaporativeLightScattering Detector.
Default Calculation: If EvaporativeLightScattering is selected, automatically set to the first entry in NebulizerGas.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardNebulizerGasHeating
Indicates if the sheath gas that carries samples in the EvaporativeLightScattering Detector is heated for Standard measurement.
Default Calculation: If EvaporativeLightScattering Detector is selected and StandardNebulizerGas is True, automatically set to the first entry in NebulizerGasHeating.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardNebulizerHeatingPower
The relative magnitude of the heating element used to heat the sheath gas for the EvaporativeLightScattering Detector for Standard measurement (Corresponding temperature not measured by the manufacturer). Higher percent values correspond to percent of power going to heating coil.
Default Calculation: If EvaporativeLightScattering Detector is selected and StandardNebulizerGas is True, automatically set to the first entry in NebulizerHeatingPower.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: Standard
StandardNebulizerGasPressure
The applied pressure of sheath gas for the EvaporativeLightScattering Detector for Standard measurement (Flow rate unmeasured by the manufacturer). Higher pressure corresponds to faster sheath gas flow.
Default Calculation: If EvaporativeLightScattering Detector is selected and StandardNebulizerGas is True, automatically set to the first entry in NebulizerGasPressure.
Pattern Description: Greater than or equal to 20 pounds
‐force per inch squared and less than or equal to 60 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[20*PSI, 60*PSI] | Automatic) | Null
Index Matches to: Standard
StandardDriftTubeTemperature
The set temperature of the chamber thermostat through which the nebulized analytes flow within the EvaporativeLightScattering Detector for Standard samples. The purpose to heat the drift tube is to evaporate any unevaporated solvent remaining in the flow from the nebulizer.
Default Calculation: If EvaporativeLightScattering Detector is selected and StandardNebulizerGas is True, automatically set to the first entry in DriftTubeTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 100*Celsius] | Automatic) | Null
Index Matches to: Standard
StandardELSDGain
The percent of maximum voltage sent to the Photo Multiplier Tube (PMT) for signal amplification for the EvaporativeLightScattering measurement. The percentage value specified here is converted into a unitless factor from 0 to 1000 which the software accepts to modulate the voltage for the PMT.
Default Calculation: If EvaporativeLightScattering Detector is selected and StandardNebulizerGas is True, automatically set to the first entry in ELSDGain.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: Standard
StandardELSDSamplingRate
The frequency of evaporative light scattering measurement for Standard samples. Lower values will be less susceptible to noise but will record less frequently across time. Lower or higher values do not affect the y axis of the measurement.
Default Calculation: If EvaporativeLightScattering Detector is selected and NebulizerGas is True, automatically set to the first entry in ELSDSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second)] | Automatic) | Null
Index Matches to: Standard
StandardStorageCondition
The non-default conditions under which any standards used by this experiment should be stored after the protocol is completed. If left unset, Standard samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: Standard
Blanks
Blank
The object(s) (samples) to inject typically as negative controls (e.g. to test effects stemming from injection, sample solvent, impurities on the column or buffer).
Default Calculation: If any other Blank option is specified or RefractiveIndex Detector is selected, automatically set to the specified BufferA or Model[Sample, "Milli-Q water"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankInjectionVolume
The physical quantity of Blank sample that is loaded into the flow path on the selected instrument along with the mobile phase onto the stationary phase.
Default Calculation: Automatically set equal to the first entry in InjectionVolume.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 16 milliliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 16*Milliliter] | Automatic) | Null
BlankFrequency
The frequency at which Blank measurements will be inserted between Sample.
Default Calculation: If injectionTable is given, automatically set to Null and the sequence of Blanks specified in the InjectionTable will be used in the experiment. If any other Blank option is specified, automatically set to FirstAndLast.
Pattern Description: Greater than 0 in increments of 1 or None, First, Last, FirstAndLast, or GradientChange or Null.
Programmatic Pattern: (((None | First | Last | FirstAndLast | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
BlankColumnPosition
The position of the column selector valve and the desired column configuration that will be used for each blank sample as it is injected.
Default Calculation: For a batch of samples automatically set from the specified Column option. If InjectionTable is specified, set from the Column Position for blank Type injections.
Pattern Description: PositionA, PositionB, PositionC, PositionD, PositionE, PositionF, PositionG, or PositionH or Null.
Programmatic Pattern: (ColumnPositionP | Automatic) | Null
BlankColumnTemperature
The temperature of the column assembly throughout the Blank gradient and measurement.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the BlankGradient option or the column temperature for the sample in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[5*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
BlankGradientA
The composition of BufferA within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientB, BlankGradientC, and BlankGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientB
The composition of BufferB within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientA, BlankGradientC, and BlankGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientC
The composition of BufferC within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientA, BlankGradientB, and BlankGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientD
The composition of BufferD within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientA, BlankGradientB, and BlankGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the BlankGradient options during Blank measurement. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If BlankGradient option is specified, automatically set from the method given in the BlankGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 200 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)]}..}) | Automatic) | Null
BlankGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow during Blank measurement. Specific parameters of a gradient object can be overridden by specific options. Differential Refractive Index Reference Loading refers to the HPLC refractive index loading reference values as shown in the Figure 2.7.4. When open, the flow downstream of the column is loaded into the two flow cells simultaneously.
Default Calculation: Automatically set to best meet all the BlankGradient options (e.g. BlankGradientA, BlankGradientB, BlankGradientC, BlankGradientD, BlankFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate, Differential Refractive Index Reference Loading} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)], Open | Closed | None | Automatic}..}) | Automatic) | Null
BlankAbsorbanceWavelength
For Blank measurement, the wavelength of light passed through the flow cell for the UVVis Detector. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 950*Nanometer] | All | RangeP[190*Nanometer, 950*Nanometer] ;; RangeP[200*Nanometer, 950*Nanometer]) | Automatic) | Null
BlankWavelengthResolution
The increment in wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector for Blank measurement.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 0.1 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[0.1*Nanometer, 12.*Nanometer] | Automatic) | Null
BlankUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the sample for the PhotoDiodeArray Detector for Blank measurement.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set as the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankAbsorbanceSamplingRate
The number of times the absorbance measurement is made per second during Blank measurement. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 120 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 120*(1/Second)] | Automatic) | Null
BlankSmoothingTimeConstant
The time window used on the instrument software for data filtering in absorbance collection of Blank sample, affecting baseline smoothing and peak height degradation. Raw absorbance signals within the time window are smoothed using a weighted moving average, with the result applied to the leftmost point of the window to effectively suppress high-frequency noise. Optimizing this parameter improves signal-to-noise ratio: shorter time constants give faster response and sharper peaks, while longer ones enhance sensitivity by reducing noise. For Waters instruments, the setting can be a numeric value or an enumeration - Small, Medium, or Large - corresponding to 1, 2, or 4 divided by AbsorbanceSamplingRate. Dionex instruments require a numeric value between 0.01 and 4.55 Second. Agilent instruments accept only specific values based on AbsorbanceSamplingRate. For details, please refer to Figure 3.1 in the ExperimentHPLC help file, and the SmoothingTimeConstants field in Model[Instrument, HPLC].
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is requested on a Waters HPLC instrument, automatically set to the first entry in SmoothingTimeConstant.
Pattern Description: Greater than or equal to 0.0125 seconds and less than or equal to 5 seconds or Small, Medium, or Large or Null.
Programmatic Pattern: ((HPLCSmoothingTimeConstantP | RangeP[0.0125*Second, 5*Second]) | Automatic) | Null
BlankExcitationWavelength
The wavelength(s) that is used to excite fluorescence in the Blank sample in the Fluorescence Detector.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in ExcitationWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
BlankEmissionWavelength
The wavelength(s) of light at which fluorescence emitted from the Blank sample is measured in the Fluorescence Detector.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in EmissionWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
BlankEmissionCutOffFilter
The cut-off wavelength to pre-select the emitted light from the Blank sample and allow only the light with wavelength above the desired value to pass, before the light enters emission monochromator for final wavelength selection for Ultimate 3000 with FLR Detector. If set to None, no cut-off filter is used.
Default Calculation: If a Fluorescence Detector with a cut-off filter wheel is selected, automatically set to the first entry in EmissionCutOffFilter.
Pattern Description: 280 nanometers, 370 nanometers, 435 nanometers, 530 nanometers, or None or Null.
Programmatic Pattern: (HPLCEmissionCutOffFilterP | Automatic) | Null
BlankFluorescenceGain
For each BlankExcitationWavelength/BlankEmissionWavelength pair, the signal amplification factor which modulates the percentage of maximum voltage that can be applied to the Photomultiplier Tube of the Fluorescence Detector during Standard measurement. Linear increase in voltage applied to the Photomultiplier tube leads to an exponential change in RFU signal. Variable Fluorescence Sensitivity implies a different fluorescence sensitivity for each Excitation/Emission Wavelength pair.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in FluorescenceGain.
Pattern Description: Constant or Variable Fluorescence Sensitivity or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {RangeP[0*Percent, 100*Percent]..}) | Automatic) | Null
BlankFluorescenceFlowCellTemperature
The temperature that the thermostat inside the fluorescence flow cell of the Fluorescence Detector is set to during the fluorescence measurement of the Blank sample.
Default Calculation: If Fluorescence Detector is selected and temperature control is available on that unit, automatically set to the first entry in FluorescenceFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[25*Celsius, 50*Celsius] | Ambient) | Automatic) | Null
BlankLightScatteringLaserPower
The laser power filter used in the Multi-Angle static Light Scattering (MALS) and Dynamic Light Scattering (DLS) Detector for the measurement of the Blank sample. 100% means that no filter is being used to restrict the laser power.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to the first entry in LightScatteringLaserPower.
Pattern Description: Greater than or equal to 10 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[10*Percent, 100*Percent] | Automatic) | Null
BlankLightScatteringFlowCellTemperature
The temperature that the thermostat inside the flow cell of the Detector is set to during the Multi-Angle static Light Scattering (MALS) and/or Dynamic Light Scattering (DLS) measurement of the Blank sample.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to the first entry in LightScatteringFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to -15 degrees Celsius and less than or equal to 210 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[-15*Celsius, 210*Celsius] | Ambient) | Automatic) | Null
BlankRefractiveIndexMethod
The type of refractive index measurement of the Refractive Index (RI) Detector for the measurement of the Blank. When DifferentialRefractiveIndex is selected, the refractive index difference between the flow downstream sample and the reference solvent is measured. See Figure 2.7.4 for more information.
Default Calculation: If RefractiveIndex Detector is selected and Differential Refractive Index Reference Loading is set to Closed in BlankGradient, automatically set to DifferentialRefractiveIndex. Otherwise automatically set to RefractiveIndex.
Pattern Description: RefractiveIndex or DifferentialRefractiveIndex or Null.
Programmatic Pattern: ((RefractiveIndex | DifferentialRefractiveIndex) | Automatic) | Null
BlankRefractiveIndexFlowCellTemperature
The temperature that the thermostat inside the refractive index flow cell of the Refractive Index (RI) Detector is set to during the refractive index measurement of the Blank sample.
Default Calculation: If RefractiveIndex Detector is selected, automatically set to the first entry in RefractiveIndexFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 65 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[4*Celsius, 65*Celsius] | Ambient) | Automatic) | Null
BlankNebulizerGas
Indicates if Nitrogen sheath gas is flowed along with the Blank sample within the EvaporativeLightScattering Detector.
Default Calculation: If EvaporativeLightScattering is selected, automatically set to the first entry in NebulizerGas.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankNebulizerGasHeating
Indicates if the sheath gas that carries samples in the EvaporativeLightScattering Detector is heated for Blank measurement.
Default Calculation: If EvaporativeLightScattering Detector is selected and BlankNebulizerGas is True, automatically set to the first entry in NebulizerGasHeating.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankNebulizerHeatingPower
The relative magnitude of the heating element used to heat the sheath gas for the EvaporativeLightScattering Detector for Blank measurement (Corresponding temperature not measured by the manufacturer). Higher percent values correspond to percent of power going to heating coil.
Default Calculation: If EvaporativeLightScattering Detector is selected and BlankNebulizerGas is True, automatically set to the first entry in NebulizerHeatingPower.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
BlankNebulizerGasPressure
The applied pressure of sheath gas for the EvaporativeLightScattering Detector for Blank measurement (Flow rate unmeasured by the manufacturer). Higher pressure corresponds to faster sheath gas flow.
Default Calculation: If EvaporativeLightScattering Detector is selected and BlankNebulizerGas is True, automatically set to the first entry in NebulizerGasPressure.
Pattern Description: Greater than or equal to 20 pounds
‐force per inch squared and less than or equal to 60 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[20*PSI, 60*PSI] | Automatic) | Null
BlankDriftTubeTemperature
The set temperature of the chamber thermostat through which the nebulized analytes flow within the EvaporativeLightScattering Detector for Blank samples. The purpose to heat the drift tube is to evaporate any unevaporated solvent remaining in the flow from the nebulizer.
Default Calculation: If EvaporativeLightScattering Detector is selected and BlankNebulizerGas is True, automatically set to the first entry in DriftTubeTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 100*Celsius] | Automatic) | Null
BlankELSDGain
The percent of maximum voltage sent to the Photo Multiplier Tube (PMT) for signal amplification for the EvaporativeLightScattering measurement. The percentage value specified here is converted into a unitless factor from 0 to 1000 which the software accepts to modulate the voltage for the PMT.
Default Calculation: If EvaporativeLightScattering Detector is selected and BlankNebulizerGas is True, automatically set to the first entry in ELSDGain.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
BlankELSDSamplingRate
The frequency of evaporative light scattering measurement for Blank Samples. Lower values will be less susceptible to noise but will record less frequently across time. Lower or higher values do not affect the y axis of the measurement.
Default Calculation: If EvaporativeLightScattering Detector is selected and BlankNebulizerGas is True, automatically set to the first entry in ELSDSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second)] | Automatic) | Null
BlankStorageCondition
The non-default conditions under which any blanks used by this experiment should be stored after the protocol is completed. If left unset, Blank samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Column Prime
ColumnRefreshFrequency
The frequency of column prime inserted into the order of analyte injections at which solvent is flowed to equilibrate the column in order to remove contaminants and reset the gradient to match the starting percentage of the subsequent injection. An initial column prime and final column flush will be performed unless Null or None is specified. For First, it is performed at the beginning. For Last, it is performed at the end. For FirstAndLast, it is performed both at the beginning and end. For GradientChange, it is performed every time a change in the gradient is encountered for the injections. A Number indicates the number of injections after which it is performed and also in the beginning (eg: for 2, it is performed at the start and after 2nd, 4th, 6th and so on injections).
Default Calculation: Automatically set to Null when InjectionTable option is specified (meaning that this option is inconsequential) or no column is used in the experiment; otherwise, set to GradientChange.
Pattern Description: Greater than 0 in increments of 1 or None, FirstAndLast, First, Last, or GradientChange or Null.
Programmatic Pattern: (((None | FirstAndLast | First | Last | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeTemperature
The column assembly's temperature at which the ColumnPrimeGradient is run.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the ColumnPrimeGradient option or the column temperature for the column prime in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[5*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeGradientA
The composition of BufferA within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientB, ColumnPrimeGradientC, and ColumnPrimeGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeGradientB
The composition of BufferB within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientA, ColumnPrimeGradientC, and ColumnPrimeGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeGradientC
The composition of BufferC within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientA, ColumnPrimeGradientB, and ColumnPrimeGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeGradientD
The composition of BufferD within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientA, ColumnPrimeGradientB, and ColumnPrimeGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the ColumnPrimeGradient options during column prime. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If ColumnPrimeGradient option is specified, automatically set from the method given in the ColumnPrimeGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 200 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow for column prime. Specific parameters of a gradient object can be overridden by specific options. Differential Refractive Index Reference Loading refers to the HPLC refractive index loading reference values as shown in the Figure 2.7.4. When open, the flow downstream of the column is loaded into the two flow cells simultaneously.
Default Calculation: Automatically set to best meet all the ColumnPrimeGradient options (e.g. ColumnPrimeGradientA, ColumnPrimeGradientB, ColumnPrimeGradientC, ColumnPrimeGradientD, ColumnPrimeFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate, Differential Refractive Index Reference Loading} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)], Open | Closed | None | Automatic}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeAbsorbanceWavelength
The wavelength of light passed through the flow for the UVVis and Detector for detection during column prime. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the same value as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 950*Nanometer] | All | RangeP[190*Nanometer, 950*Nanometer] ;; RangeP[200*Nanometer, 950*Nanometer]) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeWavelengthResolution
The increment in wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector during column prime.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the same value as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 0.1 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[0.1*Nanometer, 12.*Nanometer] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the flow for PhotoDiodeArray Detector during column prime.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set to the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeAbsorbanceSamplingRate
The number of times an absorbance measurement is made per second during column prime. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 120 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 120*(1/Second)] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeSmoothingTimeConstant
The time window used on the instrument software for data filtering in absorbance collection of column prime, affecting baseline smoothing and peak height degradation. Raw absorbance signals within the time window are smoothed using a weighted moving average, with the result applied to the leftmost point of the window to effectively suppress high-frequency noise. Optimizing this parameter improves signal-to-noise ratio: shorter time constants give faster response and sharper peaks, while longer ones enhance sensitivity by reducing noise. For Waters instruments, the setting can be a numeric value or an enumeration - Small, Medium, or Large - corresponding to 1, 2, or 4 divided by AbsorbanceSamplingRate. Dionex instruments require a numeric value between 0.01 and 4.55 Second. Agilent instruments accept only specific values based on AbsorbanceSamplingRate. For details, please refer to Figure 3.1 in the ExperimentHPLC help file, and the SmoothingTimeConstants field in Model[Instrument, HPLC].
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is requested on a Waters HPLC instrument, automatically set to the first entry in SmoothingTimeConstant.
Pattern Description: Greater than or equal to 0.0125 seconds and less than or equal to 5 seconds or Small, Medium, or Large or Null.
Programmatic Pattern: ((HPLCSmoothingTimeConstantP | RangeP[0.0125*Second, 5*Second]) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeExcitationWavelength
The wavelength(s) of light that is used to excite fluorescence in the Fluorescence Detector during column prime.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in ExcitationWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeEmissionWavelength
The wavelength(s) of light at which fluorescence emitted from the flow downstream of the column is measured in the Fluorescence Detector during column prime.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in EmissionWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeEmissionCutOffFilter
The cut-off wavelength to pre-select the emitted light from the flow downstream of the column and allow only the light with wavelength above the desired value to pass, before the light enters emission monochromator for final wavelength selection during column prime for Ultimate 3000 with FLR Detector. If set to None, no cut-off filter is used.
Default Calculation: If a Fluorescence Detector with a cut-off filter wheel is selected, automatically set to the first entry in EmissionCutOffFilter.
Pattern Description: 280 nanometers, 370 nanometers, 435 nanometers, 530 nanometers, or None or Null.
Programmatic Pattern: (HPLCEmissionCutOffFilterP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeFluorescenceGain
For each ColumnPrimeExcitationWavelength/ColumnPrimeEmissionWavelength pair, the signal amplification factor which modulates the percentage of maximum voltage that can be applied to the Photomultiplier Tube of the Fluorescence Detector during column prime. Linear increase in voltage applied to the Photomultiplier tube leads to an exponential change in RFU signal. Variable Fluorescence Sensitivity implies a different fluorescence sensitivity for each Excitation/Emission Wavelength pair.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in FluorescenceGain.
Pattern Description: Constant or Variable Fluorescence Sensitivity or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {RangeP[0*Percent, 100*Percent]..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeFluorescenceFlowCellTemperature
The temperature that the thermostat inside the fluorescence flow cell of the Fluorescence Detector is set to during column prime.
Default Calculation: If Fluorescence Detector is selected and temperature control is available on that unit, automatically set to the first entry in FluorescenceFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[25*Celsius, 50*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeLightScatteringLaserPower
The laser power filter used in the Multi-Angle static Light Scattering (MALS) and Dynamic Light Scattering (DLS) Detector during column prime measurement. 100% means that no filter is being used to restrict the laser power.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to the first entry in LightScatteringLaserPower.
Pattern Description: Greater than or equal to 10 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[10*Percent, 100*Percent] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeLightScatteringFlowCellTemperature
The temperature that the thermostat inside the flow cell inside the Multi-Angle static Light Scattering (MALS) and Dynamic Light Scattering (DLS) detector is set to during column prime.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to the first entry in LightScatteringFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to -15 degrees Celsius and less than or equal to 210 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[-15*Celsius, 210*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeRefractiveIndexMethod
The type of refractive index measurement of the Refractive Index (RI) Detector during column prime. When DifferentialRefractiveIndex is selected, the refractive index difference between the flow downstream sample and the reference solvent is measured. See Figure 2.7.4 for more information.
Default Calculation: If RefractiveIndex Detector is selected and Differential Refractive Index Reference Loading is set to Closed in ColumnPrimeGradient, automatically set to DifferentialRefractiveIndex. Otherwise automatically set to RefractiveIndex.
Pattern Description: RefractiveIndex or DifferentialRefractiveIndex or Null.
Programmatic Pattern: ((RefractiveIndex | DifferentialRefractiveIndex) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeRefractiveIndexFlowCellTemperature
The temperature that the thermostat inside the refractive index flow cell of the Refractive Index (RI) Detector is set to during column prime.
Default Calculation: If RefractiveIndex Detector is selected, automatically set to the first entry in RefractiveIndexFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 65 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[4*Celsius, 65*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeNebulizerGas
Indicates if Nitrogen sheath gas is flowed with the buffer(s) within the EvaporativeLightScattering Detector during column prime.
Default Calculation: If EvaporativeLightScattering is selected, automatically set to the first entry in NebulizerGas.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeNebulizerGasHeating
Indicates if the sheath gas that carries buffer(s) in the EvaporativeLightScattering Detector is heated during column prime.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnPrimeNebulizerGas is True, automatically set to the first entry in NebulizerGasHeating.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeNebulizerHeatingPower
The relative magnitude of the heating element used to heat the sheath gas for the EvaporativeLightScattering Detector during column prime (Corresponding temperature not measured by the manufacturer). Higher percent values correspond to percent of power going to heating coil.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnPrimeNebulizerGas is True, automatically set to the first entry in NebulizerHeatingPower.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeNebulizerGasPressure
The applied pressure of sheath gas for the EvaporativeLightScattering Detector during column prime (Flow rate unmeasured by the manufacturer). Higher pressure corresponds to faster sheath gas flow.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnPrimeNebulizerGas is True, automatically set to the first entry in NebulizerGasPressure.
Pattern Description: Greater than or equal to 20 pounds
‐force per inch squared and less than or equal to 60 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[20*PSI, 60*PSI] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeDriftTubeTemperature
The set temperature of the chamber thermostat through which the nebulized analytes flow within the EvaporativeLightScattering Detector during Column Prime. The purpose to heat the drift tube is to evaporate any unevaporated solvent remaining in the flow from the nebulizer.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnPrimeNebulizerGas is True, automatically set to the first entry in DriftTubeTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 100*Celsius] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeELSDGain
The percent of maximum voltage sent to the Photo Multiplier Tube (PMT) for signal amplification for the EvaporativeLightScattering measurement. The percentage value specified here is converted into a unitless factor from 0 to 1000 which the software accepts to modulate the voltage for the PMT.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnPrimeNebulizerGas is True, automatically set to the first entry in ELSDGain.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnPrimeELSDSamplingRate
The frequency of evaporative light scattering measurement during column prime. Lower values will be less susceptible to noise but will record less frequently across time. Lower or higher values do not affect the y axis of the measurement.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnPrimeNebulizerGas is True, automatically set to the first entry in ELSDSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second)] | Automatic) | Null
Index Matches to: ColumnSelector
Column Flush
ColumnFlushTemperature
The column assembly's temperature at which the ColumnFlushGradient is run.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the ColumnFlushGradient option or the column temperature for the column flush in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[5*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushGradientA
The composition of BufferA within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientB, ColumnFlushGradientC, and ColumnFlushGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushGradientB
The composition of BufferB within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientA, ColumnFlushGradientC, and ColumnFlushGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushGradientC
The composition of BufferC within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientA, ColumnFlushGradientB, and ColumnFlushGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushGradientD
The composition of BufferD within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientA, ColumnFlushGradientB, and ColumnFlushGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the ColumnFlushGradient options during column flush. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If ColumnFlushGradient option is specified, automatically set from the method given in the ColumnFlushGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 200 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)]}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow for column prime. Specific parameters of a gradient object can be overridden by specific options. Differential Refractive Index Reference Loading refers to the HPLC refractive index loading reference values as shown in the Figure 2.7.4. When open, the flow downstream of the column is loaded into the two flow cells simultaneously.
Default Calculation: Automatically set to best meet the values specified in ColumnFlushGradient options (e.g. ColumnFlushGradientA, ColumnFlushGradientB, ColumnFlushGradientC, ColumnFlushGradientD, ColumnFlushFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate, Differential Refractive Index Reference Loading} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 200*(Milliliter/Minute)], Open | Closed | None | Automatic}..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushAbsorbanceWavelength
The wavelength of light passed through the flow for the UVVis and Detector for detection during column flush. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the same value as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 950*Nanometer] | All | RangeP[190*Nanometer, 950*Nanometer] ;; RangeP[200*Nanometer, 950*Nanometer]) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushWavelengthResolution
The increment of wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector during column flush.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the same value as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 0.1 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[0.1*Nanometer, 12.*Nanometer] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the flow for PhotoDiodeArray Detector during column flush.
Default Calculation: If a PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set to the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushAbsorbanceSamplingRate
The number of times an absorbance measurement is made per second during column flush. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is selected or available on the selected instrument, automatically set equal to the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 120 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 120*(1/Second)] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushSmoothingTimeConstant
The time window used on the instrument software for data filtering in absorbance collection of column flush, affecting baseline smoothing and peak height degradation. Raw absorbance signals within the time window are smoothed using a weighted moving average, with the result applied to the leftmost point of the window to effectively suppress high-frequency noise. Optimizing this parameter improves signal-to-noise ratio: shorter time constants give faster response and sharper peaks, while longer ones enhance sensitivity by reducing noise. For Waters instruments, the setting can be a numeric value or an enumeration - Small, Medium, or Large - corresponding to 1, 2, or 4 divided by AbsorbanceSamplingRate. Dionex instruments require a numeric value between 0.01 and 4.55 Second. Agilent instruments accept only specific values based on AbsorbanceSamplingRate. For details, please refer to Figure 3.1 in the ExperimentHPLC help file, and the SmoothingTimeConstants field in Model[Instrument, HPLC].
Default Calculation: If a UVVis Detector or PhotoDiodeArray Detector is requested on a Waters HPLC instrument, automatically set to the first entry in SmoothingTimeConstant.
Pattern Description: Greater than or equal to 0.0125 seconds and less than or equal to 5 seconds or Small, Medium, or Large or Null.
Programmatic Pattern: ((HPLCSmoothingTimeConstantP | RangeP[0.0125*Second, 5*Second]) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushExcitationWavelength
The wavelength(s) of light that is used to excite fluorescence in the Fluorescence Detector during column flush.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in ExcitationWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushEmissionWavelength
The wavelength(s) of light at which fluorescence emitted from the flow downstream of the column is measured in the Fluorescence Detector during column flush.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in EmissionWavelength.
Pattern Description: Multi-Channel or Single-Channel or Null.
Programmatic Pattern: ((RangeP[200*Nanometer, 1200*Nanometer] | {RangeP[200*Nanometer, 890*Nanometer]..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushEmissionCutOffFilter
The cut-off wavelength to pre-select the emitted light from the flow downstream of the column and allow only the light with wavelength above the desired value to pass, before the light enters emission monochromator for final wavelength selection during column flush for Ultimate 3000 with FLR Detector. If set to None, no cut-off filter is used.
Default Calculation: If a Fluorescence Detector with a cut-off filter wheel is selected, automatically set to the first entry in EmissionCutOffFilter.
Pattern Description: 280 nanometers, 370 nanometers, 435 nanometers, 530 nanometers, or None or Null.
Programmatic Pattern: (HPLCEmissionCutOffFilterP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushFluorescenceGain
For each ColumnFlushExcitationWavelength/ColumnFlushEmissionWavelength pair, the signal amplification factor which modulates the percentage of maximum voltage that can be applied to the Photomultiplier Tube of the Fluorescence Detector during column flush. Linear increase in voltage applied to the Photomultiplier tube leads to an exponential change in RFU signal. Variable Fluorescence Sensitivity implies a different fluorescence sensitivity for each Excitation/Emission Wavelength pair.
Default Calculation: If Fluorescence Detector is selected, automatically set to the first entry in FluorescenceGain.
Pattern Description: Constant or Variable Fluorescence Sensitivity or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {RangeP[0*Percent, 100*Percent]..}) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushFluorescenceFlowCellTemperature
The temperature that the thermostat inside the fluorescence flow cell of the Fluorescence Detector is set to during column flush.
Default Calculation: If Fluorescence Detector is selected and temperature control is available on that unit, automatically set to the first entry in FluorescenceFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[25*Celsius, 50*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushLightScatteringLaserPower
The laser power filter used in the Multi-Angle static Light Scattering (MALS) and Dynamic Light Scattering (DLS) Detector during column flush measurement. 100% means that no filter is being used to restrict the laser power.
Default Calculation: If MultiAngleLightScattering Detector and/or DynamicLightScattering Detector are selected, automatically set to the first entry in LightScatteringLaserPower.
Pattern Description: Greater than or equal to 10 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[10*Percent, 100*Percent] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushLightScatteringFlowCellTemperature
The temperature that the thermostat inside the flow cell inside the Multi-Angle static Light Scattering (MALS) and Dynamic Light Scattering (DLS) detector is set to during column flush.
Default Calculation: If MultiAngleLightScattering detector and/or DynamicLightScattering detector are selected, automatically set to the first entry in LightScatteringFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to -15 degrees Celsius and less than or equal to 210 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[-15*Celsius, 210*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushRefractiveIndexMethod
The type of refractive index measurement of the Refractive Index (RI) Detector during column flush. When DifferentialRefractiveIndex is selected, the refractive index difference between the flow downstream sample and the reference solvent is measured. See Figure 2.7.4 for more information.
Default Calculation: If RefractiveIndex Detector is selected and Differential Refractive Index Reference Loading is set to Closed in ColumnFlushGradient, automatically set to DifferentialRefractiveIndex. Otherwise automatically set to RefractiveIndex.
Pattern Description: RefractiveIndex or DifferentialRefractiveIndex or Null.
Programmatic Pattern: ((RefractiveIndex | DifferentialRefractiveIndex) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushRefractiveIndexFlowCellTemperature
The temperature that the thermostat inside the refractive index flow cell of the Refractive Index (RI) Detector is set to during column flush.
Default Calculation: If RefractiveIndex detector is selected, automatically set to the first entry in RefractiveIndexFlowCellTemperature.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 65 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[4*Celsius, 65*Celsius] | Ambient) | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushNebulizerGas
Indicates if Nitrogen sheath gas is flowed with the buffer(s) within the EvaporativeLightScattering Detector during column flush.
Default Calculation: If EvaporativeLightScattering is selected, automatically set to the first entry in NebulizerGas.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushNebulizerGasHeating
Indicates if the sheath gas that carries buffer(s) in the EvaporativeLightScattering Detector is heated during column flush.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnFlushNebulizerGas is True, automatically set to the first entry in NebulizerGasHeating.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushNebulizerHeatingPower
The relative magnitude of the heating element used to heat the sheath gas for the EvaporativeLightScattering Detector during column flush (Corresponding temperature not measured by the manufacturer). Higher percent values correspond to percent of power going to heating coil.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnFlushNebulizerGas is True, automatically set to the first entry in NebulizerHeatingPower.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushNebulizerGasPressure
The applied pressure of sheath gas for the EvaporativeLightScattering Detector during column flush (Flow rate unmeasured by the manufacturer). Higher pressure corresponds to faster sheath gas flow.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnFlushNebulizerGas is True, automatically set to the first entry in NebulizerGasPressure.
Pattern Description: Greater than or equal to 20 pounds
‐force per inch squared and less than or equal to 60 pounds
‐force per inch squared or Null.
Programmatic Pattern: (RangeP[20*PSI, 60*PSI] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushDriftTubeTemperature
The set temperature of the chamber thermostat through which the nebulized analytes flow within the EvaporativeLightScattering Detector during Column Flush. The purpose to heat the drift tube is to evaporate any unevaporated solvent remaining in the flow from the nebulizer.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnFlushNebulizerGas is True, automatically set to the first entry in DriftTubeTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 100*Celsius] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushELSDGain
The percent of maximum voltage sent to the Photo Multiplier Tube (PMT) for signal amplification for the EvaporativeLightScattering measurement. The percentage value specified here is converted into a unitless factor from 0 to 1000 which the software accepts to modulate the voltage for the PMT.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnFlushNebulizerGas is True, automatically set to the first entry in ELSDGain.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | Automatic) | Null
Index Matches to: ColumnSelector
ColumnFlushELSDSamplingRate
The frequency of evaporative light scattering measurement during column flush. Lower values will be less susceptible to noise but will record less frequently across time. Lower or higher values do not affect the y axis of the measurement.
Default Calculation: If EvaporativeLightScattering Detector is selected and ColumnFlushNebulizerGas is True, automatically set to the first entry in ELSDSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second)] | Automatic) | Null
Index Matches to: ColumnSelector
Model Input
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((Null | (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All)) | Automatic) | Null
Index Matches to: experiment samples
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise, automatically set to Model[Container, Plate, "96-well 2mL Deep Well Plate"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: ((Null | ObjectP[Model[Container]]) | Automatic) | Null
Index Matches to: experiment samples
Post Experiment
InjectionSampleVolumeMeasurement
Indicates if any liquid samples prepared in the sub SamplePreparationProtocols of are volume checked prior to injection to provide QC data.
Pattern Description: True or False or Null.
Programmatic Pattern: BooleanP | Null
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples