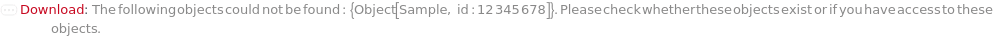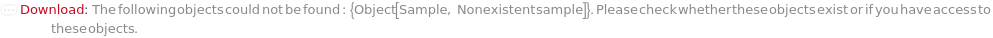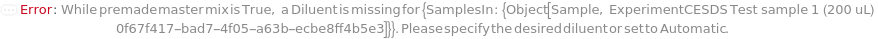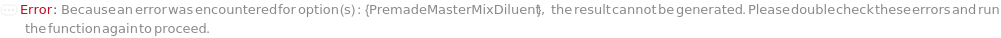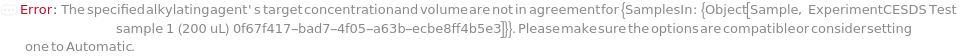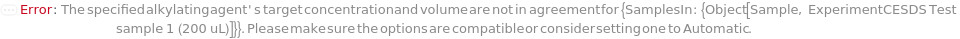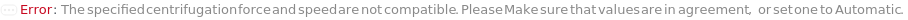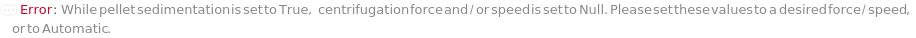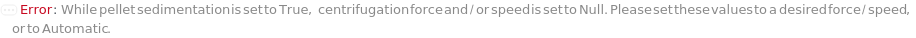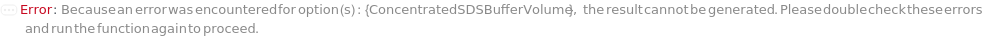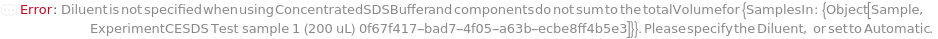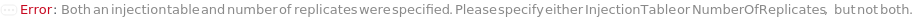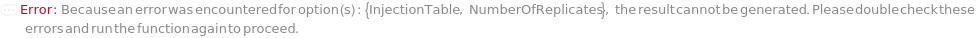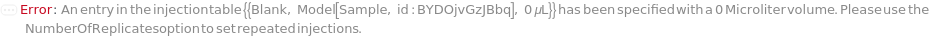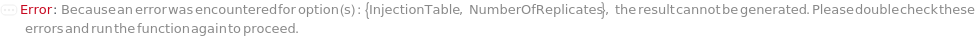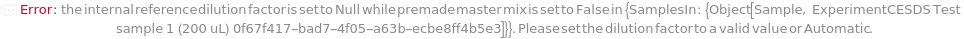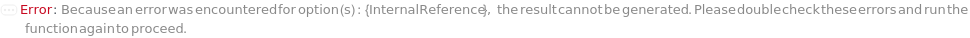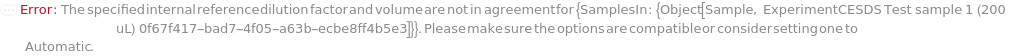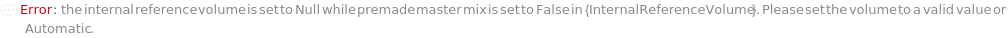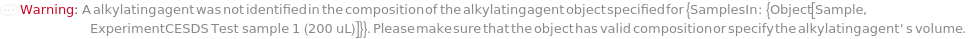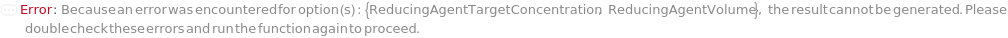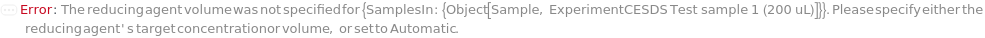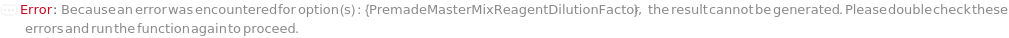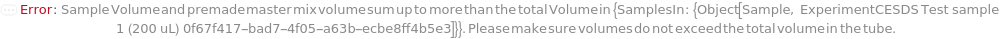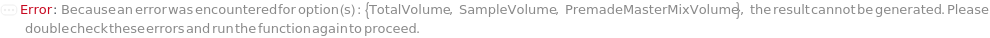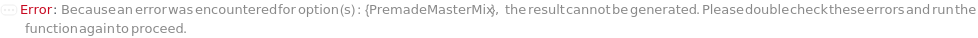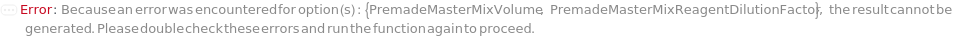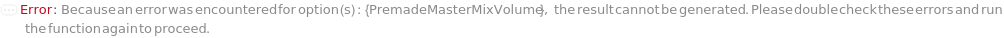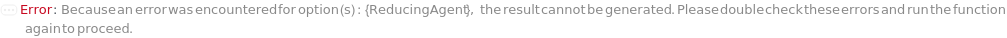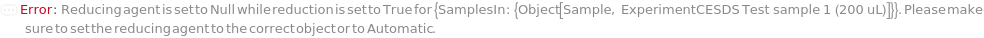General
Instrument
The capillary electrophoresis instrument that will be used by the protocol. The instrument accepts the cartridge loaded with running buffer and sequentially loads samples and resolves proteins by their size by electrophoresis through a separation matrix.
Default Value: Model[Instrument, ProteinCapillaryElectrophoresis, Maurice]
Pattern Description: An object of type or subtype Model[Instrument, ProteinCapillaryElectrophoresis] or Object[Instrument, ProteinCapillaryElectrophoresis]
Programmatic Pattern: ObjectP[{Model[Instrument, ProteinCapillaryElectrophoresis], Object[Instrument, ProteinCapillaryElectrophoresis]}]
Cartridge
The capillary electrophoresis cartridge loaded on the instrument for Capillary gel Electrophoresis-SDS (CESDS) experiments. The cartridge holds a single capillary and the anode's running buffer. CESDS cartridges can run 100 injections in up to 25 batches.
Default Value: Model[Container, ProteinCapillaryElectrophoresisCartridge, CESDS-Plus]
Pattern Description: An object of type or subtype Model[Container, ProteinCapillaryElectrophoresisCartridge] or Object[Container, ProteinCapillaryElectrophoresisCartridge] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Container, ProteinCapillaryElectrophoresisCartridge], Object[Container, ProteinCapillaryElectrophoresisCartridge]}] | _String
PurgeCartridge
Indicates if a Cartridge purge be performed before running the experiment unless there is a single batch left on the cartridge (uses an extra consumable and would count as an extra batch towards the 25 batch limit). When a cartridge has not been used for over 3 months a purge is highly recommended.
Default Value: IfRequired
Pattern Description: True, False, or IfRequired.
Programmatic Pattern: True | False | IfRequired
SampleTemperature
The temperature at which samples are maintained between sample preparation steps (SDS/reference/reagent mixing and optional denaturing) and while awaiting injection.
Pattern Description: Ambient, 4 degrees Celsius, 10 degrees Celsius, or 15 degrees Celsius.
Programmatic Pattern: Ambient | 4*Celsius | 10*Celsius | 15*Celsius
InjectionTable
The order of sample, Ladder, Standard, and Blank sample loading into the Instrument during measurement.
Default Calculation: Determined to the order of input samples articulated. Ladder, Standard and Blank samples are inserted based on the determination of LadderFrequency, StandardFrequency and BlankFrequency. For example, StandardFrequency -> FirstAndLast and BlankFrequency -> Null result in Standard samples injected first, then samples, and then the Standard sample set again at the end. A ladder sample will be loaded for every unique set of conditions (MasterMix and separation conditions) specified for SamplesIn, Standards, and Blanks, in replicates as determined by LadderFrequency.
Pattern Description: List of one or more {Type, Sample, Volume} entries.
Programmatic Pattern: {{Sample | Ladder | Standard | Blank, ObjectP[{Model[Sample], Object[Sample]}] | _String, RangeP[0*Microliter, 200*Microliter] | Automatic}..} | Automatic
NumberOfReplicates
The number of times each sample will be injected. By default, this option means technical replicates that are injected from the same position on the assay plate.
Pattern Description: Greater than or equal to 1 in increments of 1 or Null.
Programmatic Pattern: (GreaterEqualP[1, 1] | Automatic) | Null
Instrument Preparation
ConditioningAcid
The Conditioning Acid solution used to wash the capillary (every 12 injections).
Default Value: Model[Sample, CESDS Conditioning Acid]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
ConditioningBase
The Conditioning Base solution used to wash the capillary (every 12 injections).
Default Value: Model[Sample, CESDS Conditioning Base]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
ConditioningWashSolution
The solution used to wash the capillary (every 12 injections).
Default Value: Model[Sample, Milli-Q water]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
SeparationMatrix
The sieving matrix loaded onto the capillary between samples for separation. The mash-like matrix slows the migration of proteins based on their size.
Default Value: Model[Sample, CESDS Separation Matrix]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
SystemWashSolution
The solution used to wash the capillary after conditioning and, separately, rinse the tip before every injection.
Default Value: Model[Sample, CESDS Wash Solution]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
ConditioningAcidStorageCondition
The non-default storage condition for ConditioningAcid of this experiment after the protocol is completed. If left unset, samples will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
ConditioningBaseStorageCondition
The non-default storage condition for ConditioningBase of this experiment after the protocol is completed. If left unset, samples will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
SeparationMatrixStorageCondition
The non-default storage condition for SeparationMatrix of this experiment after the protocol is completed. If left unset, samples will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
SystemWashSolutionStorageCondition
The non-default storage condition for SystemWashSolution of this experiment after the protocol is completed. If left unset, samples will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
RunningBufferStorageCondition
The non-default storage condition for Bottom RunningBuffer of this experiment after the protocol is completed. If left unset, Bottom RunningBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Denaturation
Denature
Indicates if heat denaturation should be applied to all samples.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
DenaturingTemperature
The temperature to which samples will be heated to in order to linearize proteins.
Default Calculation: When Denature is True and DenaturingTemperature is set to Automatic, it will be set to 70 celsius. Alternatively, if Denature is False, it will be set to Null.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 100 degrees Celsius or Null.
Programmatic Pattern: (RangeP[$AmbientTemperature, 100*Celsius] | Automatic) | Null
DenaturingTime
The duration samples should be incubated at the DenaturingTemperature.
Default Calculation: When Denature is True and DenaturingTime is set to Automatic, it will be set to 10 minutes. Alternatively, if Denature is False, it will be set to Null.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Second, $MaxExperimentTime] | Automatic) | Null
CoolingTemperature
The temperature to which samples will be cooled to after denaturation.
Default Calculation: When Denature is True and CoolingTemperature is set to Automatic, it will be set to 4 celsius. Alternatively, if Denature is False, it will be set to Null.
Pattern Description: Greater than or equal to 0 degrees Celsius and less than or equal to 25 degrees Celsius or Null.
Programmatic Pattern: (RangeP[0*Celsius, $AmbientTemperature] | Automatic) | Null
CoolingTime
The duration samples should be incubated at the CoolingTemperature.
Default Calculation: When Denature is True and CoolingTime is set to Automatic, it will be set to 5 minutes. Alternatively, if Denature is False, it will be set to Null.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Second, $MaxExperimentTime] | Automatic) | Null
PelletSedimentation
Indicates if centrifugation should be applied to the sample to remove precipitates after denaturation.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
SedimentationCentrifugationSpeed
The speed to which the centrifuge is set to for sedimentation.
Default Calculation: When PelletSedimentation is True and SedimentationCentrifugationSpeed is set to Automatic, it will be calculated from force based on the rotor's radius.
Pattern Description: Greater than or equal to 0 revolutions per minute and less than or equal to 4150 revolutions per minute or Null.
Programmatic Pattern: (RangeP[0*RPM, 4150*RPM] | Automatic) | Null
SedimentationCentrifugationForce
The force to which the centrifuge is set to for sedimentation.
Default Calculation: When PelletSedimentation is True and SedimentationCentrifugationForce is set to Automatic, it will be calculated from Speed based on the rotor's radius, if speed is set to Automatic/Null, will default to 1000xg.
Pattern Description: Greater than or equal to 0 standard accelerations due to gravity on the surface of the earth and less than or equal to 3755 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: (RangeP[0*GravitationalAcceleration, 3755*GravitationalAcceleration] | Automatic) | Null
SedimentationCentrifugationTime
The duration samples should be centrifuged to pellet precipitates.
Default Value: 10 minutes
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 72 hours or Null.
Programmatic Pattern: RangeP[0*Second, $MaxExperimentTime] | Null
SedimentationCentrifugationTemperature
The temperature to which samples will be cooled to during centrifugation.
Default Value: 4 degrees Celsius
Pattern Description: Greater than or equal to 0 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
Programmatic Pattern: RangeP[0*Celsius, 40*Celsius] | Null
Sample Preparation
SampleVolume
Indicates the volume drawn from the sample to the assay tube. Each tube contains a Sample, an InternalReference, an SDSBuffer, and a ReducingAgent and/or an AlkylatingAgent.
Default Calculation: When SampleVolume is set to Automatic, the volume is calculated for the composition of the sample to reach 1 mg / ml by 1 Milligram/Milliliter * TotalVolume / Sample [Concentration]. If an injection table is available, volumes will be taken from it.
Pattern Description: Greater than 0 microliters.
Programmatic Pattern: GreaterP[0*Microliter] | Automatic
Index Matches to: experiment samples
TotalVolume
Indicates the final volume in the assay tube prior to loading onto AssayContainer. Each tube contains a Sample, an InternalReference, an SDSBuffer, and a ReducingAgent and/or an AlkylatingAgent.
Default Value: 100 microliters
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 200 microliters.
Programmatic Pattern: RangeP[50*Microliter, 200*Microliter]
Index Matches to: experiment samples
PremadeMasterMix
Indicates if samples should be mixed with PremadeMasterMix that includes an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Default Calculation: When PremadeMasterMix is set to Automatic, it will resolve to True if any of its downstream options is specified.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
PremadeMasterMixReagent
The premade master mix used for CESDS experiment, containing an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null
Index Matches to: experiment samples
PremadeMasterMixDiluent
The solution used to dilute the premade master mix used to its working concentration.
Default Calculation: When PremadeMasterMix is set to True and PremadeMasterMixDiluent is set to Automatic, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
PremadeMasterMixReagentDilutionFactor
The factor by which the premade mastermix should be diluted by in the final assay tube.
Default Calculation: When PremadeMasterMix is set to True and PremadeMasterMixReagentDilutionFactor is set to Automatic, it will be set as the ratio of the total volume to premade mastermix volume.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
Index Matches to: experiment samples
PremadeMasterMixVolume
The volume of the PremadeMasterMix required to reach its final concentration.
Default Calculation: When volume is set to Automatic, the volume is calculated by the division of TotalVolume by PremadeMasterMixReagentDilutionFactor. If PremadeMasterMix is set to False, PremadeMasterMixVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
PremadeMasterMixStorageCondition
The non-default storage condition for PremadeMasterMix of this experiment after the protocol is completed. If left unset, Diluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
InternalReference
The solution containing the reference analyte by which Relative Migration Time is normalized. By default a 10 KDa marker is used. It is highly recommended to include an internal standard in Sample Preparation.
Default Calculation: When PremadeMasterMix is set to False and InternalReference is set to Automatic, InternalReference will be set as Model[Sample, StockSolution, "Resuspended CESDS Internal Standard 25X"]. If PremadeMasterMix is set to True, InternalReference is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
InternalReferenceDilutionFactor
Marks how concentrated the internal standard is. For example, Model[Sample, StockSolution"Resuspended CESDS Internal Standard 25X"] is concentrated 25X, hence 2
µL should be added in Sample Preparation when final volume is set to 60 microliters.
Default Calculation: When PremadeMasterMix is set to False and InternalReferenceDilutionFactor is set to Automatic, InternalReferenceDilutionFactor will be set to 25. If PremadeMasterMix is set to True, InternalReferenceDilutionFactor is Null.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
Index Matches to: experiment samples
InternalReferenceVolume
The volume of the internal standard added to each sample.
Default Calculation: When InternalReferenceVolume is set to Automatic and PremadeMasterMix is set to False, the volume is calculated by the division of TotalVolume by InternalReferenceDilutionFactor. When PremadeMasterMix is set to True, InternalReferenceVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
ConcentratedSDSBuffer
The SDS Buffer used to dilute the sample. The final concentration of SDS in this assay must be equal to or greater than 0.5%.
Default Calculation: When PremadeMasterMix is set to False and ConcentratedSDSBuffer is set to Automatic, ConcentratedSDSBuffer will be set as Model[Sample,"1% SDS in 100mM Tris, pH 9.5"]. If PremadeMasterMix is set to True, ConcentratedSDSBuffer is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
ConcentratedSDSBufferDilutionFactor
Marks how concentrated the SDS buffer is. For example, a ConcentratedSDSBuffer that contains 1% SDS is concentrated 2X and will constitute half the TotalVolume.
Default Calculation: When PremadeMasterMix is set to False and ConcentratedSDSBufferDilutionFactor is set to Automatic, ConcentratedSDSBufferDilutionFactor will be set to 2. If PremadeMasterMix is set to True, ConcentratedSDSBufferDilutionFactor is Null.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
Index Matches to: experiment samples
Diluent
The solution used to dilute the ConcentratedSDSBuffer to working concentration.
Default Calculation: When PremadeMasterMix is set to False and Diluent is set to Automatic, Diluent will be set to Model[Sample,"Milli-Q water"]. If PremadeMasterMix is set to True, Diluent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
ConcentratedSDSBufferVolume
The volume of ConcentratedSDSBuffer added to each sample.
Default Calculation: When volume is set to Automatic and PremadeMasterMix is set to False, the volume is calculated by the division of TotalVolume by ConcentratedSDSBufferDilutionFactor. When PremadeMasterMix is set to True, ConcentratedSDSBufferVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SDSBuffer
The SDS Buffer used to dilute the sample. The final concentration of SDS in this assay must be equal or greater than 0.5%. If ConcentratedSDSBuffer is set to Null, SDSBuffer will be used to dilute the sample.
Default Calculation: When PremadeMasterMix is set to False ,ConcentratedSDSBuffer is set to Null, and SDSBuffer is set to Automatic, SDSBuffer will be set as Model[Sample,"1% SDS in 100mM Tris, pH 9.5"]. If PremadeMasterMix or ConcentratedSDSBuffer are set, SDSBuffer is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
SDSBufferVolume
The volume of SDSBuffer added to each sample.
Default Calculation: When SDSBufferVolume is set to Automatic and SDSBuffer is not Null, the volume is calculated by the difference between the TotalVolume and the volume in the tube that includes the Sample, InternalReference, ReducingAgent, and AlkylatingAgent.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
Reduction
Indicates if disulfide bridges should be chemically reduced in the sample.
Default Calculation: When automatic, Reduction will set to True if any of the Reduction options are not Null.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
ReducingAgent
The reducing agent used to reduce disulfide bridges in proteins to be added to the sample. for example,
β-mercaptoethanol or Dithiothreitol.
Default Calculation: When PremadeMasterMix is set to False and ReducingAgent is set to Automatic, ReducingAgent will be set to Model[Sample,"2-mercaptoethanol"]. If PremadeMasterMix is set to True, ReducingAgent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
ReducingAgentTargetConcentration
The final concentration of the reducing agent in the sample.
Default Calculation: When target concentration is set to Automatic, PremadeMasterMix is set to False, and no ReducingAgentVolume value is given, concentration is set to 650 millimolar if ReducingAgent is BME. If volume is given concentration will be calculated. If PremadeMasterMix is set to True, ReducingAgentTargetConcentration is Null
Pattern Description: Greater than 0 molar or greater than or equal to 0 VolumePercent and less than or equal to 100 VolumePercent or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | RangeP[0*VolumePercent, 100*VolumePercent]) | Automatic) | Null
Index Matches to: experiment samples
ReducingAgentVolume
The volume of the reducing agent required to reach its final concentration in the sample.
Default Calculation: When ReducingAgentVolume is set to Automatic and PremadeMasterMix is set to False, calculate the volume required to reach ReducingAgentTargetConcentration, if set. When ReducingAgentVolume is set to Automatic and ReducingAgentTargetConcentration is set to Null, ReducingAgentVolume is set to Null. When PremadeMasterMix is set to True, ReducingAgentVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
Alkylation
Indicates if Alkylation should be applied to the sample. Alkylation of free cysteines is useful in mitigating unexpected protein fragmentation and reproducibility issues.
Default Calculation: When automatic, Alkylation will set to True if any of the Reduction options are not Null.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
AlkylatingAgent
The alkylating agent to be added to the sample. For example, Iodoacetamide.
Default Calculation: When PremadeMasterMix is set to False and AlkylatingAgent is set to Automatic, AlkylatingAgent will be set to Model[Sample,"250 mM Iodoacetamide"]. If PremadeMasterMix is set to True, AlkylatingAgent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
AlkylatingAgentTargetConcentration
The final concentration of the alkylating agent in the sample.
Default Calculation: When target concentration is set to Automatic, PremadeMasterMix is set to False, and no AlkylatingAgentVolume value is given, concentration is set to 11.5 millimolar. If volume is given concentration will be calculated. If PremadeMasterMix is set to True, AlkylatingAgentTargetConcentration is Null.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: experiment samples
AlkylatingAgentVolume
The volume of the alkylating agent required to reach its final concentration in the sample.
Default Calculation: When AlkylatingAgentVolume is set to Automatic and PremadeMasterMix is set to False, calculate the volume required to reach AlkylatingAgentTargetConcentration, if set. When AlkylatingAgentVolume is set to Automatic and AlkylatingAgentTargetConcentration is set to Null, AlkylatingAgentVolume is set to Null. If PremadeMasterMix is set to True, AlkylatingAgentVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SedimentationSupernatantVolume
The volume of supernatant to transfer to the assay container from the sample tubes after denaturation and centrifugation. A minimum of 50 microliters are required for the analysis to proceed.
Default Calculation: When SedimentationSupernatantVolume is set to Automatic, 90% of the TotalVolume is drawn to avoid disturbing the pellet.
Pattern Description: Greater than or equal to 50 microliters or Null.
Programmatic Pattern: (GreaterEqualP[50*Microliter] | Automatic) | Null
Index Matches to: experiment samples
Storage Conditions
InternalReferenceStorageCondition
The non-default storage condition for InternalReference of this experiment after samples are transferred to assay tubes. If left unset, InternalReference will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
ConcentratedSDSBufferStorageCondition
The non-default storage condition for ConcentratedSDSBuffer of this experiment after samples are transferred to assay tubes. If left unset, ConcentratedSDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
DiluentStorageCondition
The non-default storage condition for Diluent of this experiment after the protocol is completed. If left unset, Diluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
SDSBufferStorageCondition
The non-default storage condition for SDSBuffer of this experiment after samples are transferred to assay tubes. If left unset, SDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
ReducingAgentStorageCondition
The non-default storage condition for ReducingAgent of this experiment after samples are transferred to assay tubes. If left unset, ReducingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
AlkylatingAgentStorageCondition
The non-default storage condition for AlkylatingAgent of this experiment after samples are transferred to assay tubes. If left unset, AlkylatingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
CartridgeStorageCondition
The non-default storage condition for the Cartridge after the protocol is completed. If left unset, Cartridge will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Separation conditions
RunningBuffer
The buffer in which the capillary docks for separation. This buffer must be compatible with the running buffer loaded on the CESDS cartridge (see: OnboardRunningBuffer field of the selected Cartridge).
Default Value: Model[Sample, CESDS Running Buffer - Bottom]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
InjectionVoltageDurationProfile
Series of voltages and durations to apply onto the capillary for injection. Supports up to 20 steps.
Default Value: {{4600 volts, 20 seconds}}
Pattern Description: List of one or more {Voltage, Time} entries.
Programmatic Pattern: {{RangeP[0*Volt, 6500*Volt], RangeP[0*Second, 200*Second]}..}
Index Matches to: experiment samples
SeparationVoltageDurationProfile
Series of voltages and durations to apply onto the capillary for separation. Supports up to 20 steps.
Default Calculation: When SeparationVoltageDurationProfile is set to Automatic, separation time will be set according to whether or not samples are reduced or not, and which Standard is used. If standard is IgG and samples are not reduced, set time to 25 minutes, otherwise set to 35 minutes. For all conditions, voltage is set to 5750 Volts.
Pattern Description: List of one or more {Voltage, Time} entries.
Programmatic Pattern: {{RangeP[0*Volt, 6500*Volt], RangeP[0*Minute, 180*Minute]}..} | Automatic
Index Matches to: experiment samples
Detection
UVDetectionWavelength
The wavelength used for signal detection. The hardware is currently only capable of detection at 220 nm.
Default Value: 220 nanometers
Pattern Description: An expression that matches the pattern: 220 Nanometer.
Programmatic Pattern: 220*Nanometer
Ladders
IncludeLadders
Indicates if mixtures of known analytes should be included in this experiment. A ladder contains a mixture of analytes of known size and Relative Migration Times and can be used to interpolate the molecular weight of unknown analytes. The ladder will be injected for every unique Separation set of conditions.
Default Calculation: If any ladder-related options are set, then this is set True; otherwise, false.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Ladders
Indicates the premade mixture of analytes of known molecular weight (MW) to include as reference for MW interpolation in this experiment.
Default Calculation: If IncludeLadder is True, Ladders is resolved to Model[Sample, "Unstained Protein Standard"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderStorageCondition
The non-default storage condition for Ladders of this experiment after the protocol is completed. If left unset, LadderStorageCondition will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
LadderAnalytes
Indicates the analytes included in ladder.
Default Calculation: If composition of Ladder is known, will populate LadderAnalytes accordingly. Otherwise, Null.
Pattern Description: List of one or more an object of type or subtype Model[Molecule] entries or Null.
Programmatic Pattern: ({ObjectP[Model[Molecule]]..} | Automatic) | Null
Index Matches to: Ladders
LadderAnalyteMolecularWeights
Indicates the molecular weights of analytes included in ladder.
Default Calculation: If composition of Ladder is known, will populate LadderAnalytesMolecularWeights accordingly. Otherwise, Null.
Pattern Description: List of one or more greater than 0 kilograms per mole entries or Null.
Programmatic Pattern: ({GreaterP[0*Kilodalton]..} | Automatic) | Null
Index Matches to: Ladders
LadderAnalyteLabels
Indicates the label of each analyte included in ladder.
Default Calculation: Will populate LadderAnalyteLabels according to LadderAnalytes, if set. Otherwise, Will populate LadderAnalyteLabels according to LadderAnalyteMolecularWeights.
Pattern Description: List of one or more a string entries or Null.
Programmatic Pattern: ({_String..} | Automatic) | Null
Index Matches to: Ladders
LadderFrequency
Indicates how many injections per permutation of ladder and unique set of separation conditions should be included in this experiment.
Default Calculation: If LadderFrequency is set to Automatic and Ladders is True, LadderFrequency will be set to FirstAndLast.
Pattern Description: _?(GreaterQ[#1, 0, 1] & ), FirstAndLast, First, or Last or Null.
Programmatic Pattern: ((GreaterP[0, 1] | FirstAndLast | First | Last) | Automatic) | Null
Index Matches to: Ladders
Ladder Preparation
LadderTotalVolume
Indicates the final volume in the assay tube prior to loading onto AssayContainer. Each tube contains a ladder, an InternalReference, and an SDSBuffer.
Default Calculation: If Automatic and Ladders is True, will populate LadderTotalVolume according to TotalVolume.
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[50*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderDilutionFactor
The factor by which the ladder should be diluted by in the final assay tube.
Default Calculation: When LadderDilutionFactor is set to Automatic, it is set to 2.5 by default.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
Index Matches to: Ladders
LadderVolume
The volume of ladder required to reach its final concentration.
Default Calculation: When volume is set to Automatic, the volume is calculated by the division of LadderTotalVolume by LadderDilutionFactor. If LadderDilutionFactor is Null, 40 Microliters will be used. If InjectionTable is informed, specified volumes will be used.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderPremadeMasterMix
Indicates if Ladders should be mixed with LadderPremadeMasterMix that includes an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Default Calculation: When LadderPremadeMasterMix is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Ladders
LadderPremadeMasterMixReagent
The premade master mix used for CESDS experiment, containing an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Default Calculation: When LadderPremadeMasterMixReagent is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderPremadeMasterMixDiluent
The solution used to dilute the premade master mix used to its working concentration.
Default Calculation: When LadderPremadeMasterMix is set to True and LadderPremadeMasterMixDiluent is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderPremadeMasterMixReagentDilutionFactor
The factor by which the premade mastermix should be diluted by in the final assay tube.
Default Calculation: When LadderPremadeMasterMix is set to True and LadderPremadeMasterMixReagentDilutionFactor is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
Index Matches to: Ladders
LadderPremadeMasterMixVolume
The volume of the PremadeMasterMix required to reach its final concentration.
Default Calculation: When volume is set to Automatic, and LadderPremadeMasterMix is set to True, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderPremadeMasterMixStorageCondition
The non-default storage condition for LadderPremadeMasterMix of this experiment after the protocol is completed. If left unset, LadderPremadeMasterMix will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
LadderInternalReference
The solution containing the reference analyte by which Relative Migration Time is normalized. By default a 10 KDa marker is used. It is highly recommended to include an internal standard in Sample Preparation.
Default Calculation: When LadderPremadeMasterMix is set to False and InternalReference is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderInternalReferenceDilutionFactor
Marks how concentrated the internal standard is. For example, Model[Sample,"CESDS Internal Reference 25X"] is concentrated 25X, hence 2
µL should be added in Sample Preparation when final volume is set to 50
µL.
Default Calculation: When LadderPremadeMasterMix is set to False and LadderInternalReferenceDilutionFactor is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
Index Matches to: Ladders
LadderInternalReferenceVolume
The volume of the internal standard added to each sample.
Default Calculation: When volume is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderConcentratedSDSBuffer
The SDS Buffer used to dilute standards. The final concentration of SDS in this assay must be equal to or greater than 0.5%.
Default Calculation: When LadderPremadeMasterMix is set to False and LadderConcentratedSDSBuffer is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderConcentratedSDSBufferDilutionFactor
Marks how concentrated the LadderConcentratedSDSBuffer is. For example, a LadderConcentratedSDSBuffer that contains 1% SDS is concentrated 2X and will constitute half the LadderTotalVolume.
Default Calculation: When LadderPremadeMasterMix is set to False and LadderConcentratedSDSBufferDilutionFactor is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
Index Matches to: Ladders
LadderDiluent
The solution used to dilute LadderConcentratedSDSBuffer to working concentration.
Default Calculation: When LadderPremadeMasterMix is set to False and Diluent is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderConcentratedSDSBufferVolume
The volume of LadderConcentratedSDSBufferVolume added to each sample.
Default Calculation: When volume is set to Automatic and LadderPremadeMasterMix is set to False, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderSDSBuffer
The SDS Buffer used to dilute the sample. The final concentration of SDS in this assay must be equal or greater than 0.5%.
Default Calculation: When SDS Buffer is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderSDSBufferVolume
The volume of SDSBuffer added to each sample.
Default Calculation: When LadderSDSBufferVolume is set to Automatic and LadderSDSBuffer is not Null, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderReduction
Indicates if disulfide bridges should be chemically reduced in the Ladder.
Default Calculation: When automatic, LadderReduction will set to True if any of the Reduction options are not Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Ladders
LadderReducingAgent
The reducing agent to be added to the Ladder. for example,
β-mercaptoethanol or Dithiothreitol.
Default Calculation: When LadderPremadeMasterMix is set to False and LadderReducingAgent is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderReducingAgentTargetConcentration
The final concentration of the reducing agent in the Ladder.
Default Calculation: When target concentration is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than 0 molar or greater than or equal to 0 VolumePercent and less than or equal to 100 VolumePercent or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | RangeP[0*VolumePercent, 100*VolumePercent]) | Automatic) | Null
Index Matches to: Ladders
LadderReducingAgentVolume
The volume of the reducing agent required to reach its final concentration in the Ladder.
Default Calculation: When LadderReducingAgentVolume is set to Automatic and LadderPremadeMasterMix is set to False, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderAlkylation
Indicates if Alkylation should be applied to the Ladder. Alkylation of free cysteines is useful in mitigating unexpected protein fragmentation and reproducibility issues.
Default Calculation: When automatic, LadderAlkylation will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Ladders
LadderAlkylatingAgent
The alkylating agent to be added to the Ladder. For example, Iodoacetamide.
Default Calculation: When LadderPremadeMasterMix is set to False and LadderAlkylatingAgent is set to Automatic it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Ladders
LadderAlkylatingAgentTargetConcentration
The final concentration of the alkylating agent in the Ladder.
Default Calculation: When target concentration is set to Automatic it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: Ladders
LadderAlkylatingAgentVolume
The volume of the alkylating agent required to reach its final concentration in the Ladder.
Default Calculation: When LadderAlkylatingAgentVolume is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderSedimentationSupernatantVolume
The volume of supernatant to transfer to the assay container from the Ladder tubes after denaturation and centrifugation. A minimum of 50
µL are required for the analysis to proceed.
Default Calculation: When SedimentationSupernatantVolume is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: Greater than or equal to 50 microliters or Null.
Programmatic Pattern: (GreaterEqualP[50*Microliter] | Automatic) | Null
Index Matches to: Ladders
LadderInternalReferenceStorageCondition
The non-default storage condition for LadderInternalReference of this experiment after the protocol is completed. If left unset, LadderInternalReferences will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
LadderConcentratedSDSBufferStorageCondition
The non-default storage condition for LadderConcentratedSDSBuffer of this experiment after the protocol is completed. If left unset, LadderConcentratedSDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
LadderDiluentStorageCondition
The non-default storage condition for LadderDiluent of this experiment after the protocol is completed. If left unset, LadderDiluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
LadderSDSBufferStorageCondition
The non-default storage condition for LadderSDSBuffer of this experiment after the protocol is completed. If left unset, LadderSDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
LadderReducingAgentStorageCondition
The non-default storage condition for LadderReducingAgent of this experiment after the protocol is completed. If left unset, LadderReducingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
LadderAlkylatingAgentStorageCondition
The non-default storage condition for LadderAlkylatingAgent of this experiment after the protocol is completed. If left unset, LadderAlkylatingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Ladders
Ladder Separation
LadderInjectionVoltageDurationProfile
Voltage profile for Injection. Supports up to 20 steps where each step is 0-200 Second, and 0-6500 volts.
Default Calculation: When LadderInjectionVoltageDurationProfile is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: ({{RangeP[0*Volt, 6500*Volt], RangeP[0*Second, 200*Second]}..} | Automatic) | Null
Index Matches to: Ladders
LadderSeparationVoltageDurationProfile
Voltage profile for Injection. Supports up to 20 steps.
Default Calculation: When LadderSeparationVoltageDurationProfile is set to Automatic, it will resolve according to each unique set of options applied to SamplesIn.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: ({{RangeP[0*Volt, 6500*Volt], RangeP[0*Minute, 180*Minute]}..} | Automatic) | Null
Index Matches to: Ladders
Standards
IncludeStandards
Indicates if standards should be included in this experiment. Standards contain identified analytes of known size and Relative Migration Times. Standards are used to both ensure reproducibility within and between Experiments and to interpolate the molecular weight of unknown analytes.
Default Calculation: If any Standard-related options are set, then this is set True; otherwise, false.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Standards
Indicates which standards to include.
Default Calculation: If IncludeStandards is True, Standards is resolved to Model[Sample, StockSolution, "Resuspended CESDS IgG Standard"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardStorageCondition
The non-default storage condition for Standards of this experiment after the protocol is completed. If left unset, StandardStorageCondition will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardFrequency
Indicates how many injections per standard should be included in this experiment. Sample, Standard, and Blank injection order are resolved according to InjectionTable.
Default Calculation: When StandardFrequency is set to Automatic and IncludeStandards is True, Set Frequency to FirstAnLast.
Pattern Description: _?(GreaterQ[#1, 0, 1] & ), FirstAndLast, First, or Last or Null.
Programmatic Pattern: ((GreaterP[0, 1] | FirstAndLast | First | Last) | Automatic) | Null
Index Matches to: Standards
Standard Preparation
StandardVolume
Indicates the volume drawn from the standard to the assay tube. Each tube contains a Standard, an InternalReference, an SDSBuffer, and a ReducingAgent and/or an AlkylatingAgent.
Default Calculation: When StandardVolume is set to Automatic and IncludeStandards is True, the volume is calculated to reach 1 milligram/milliliter in the assay tube, based on its composition, by StandardTotalVolume / Standard [Concentration]. If InjectionTable is informed, specified volumes will be used.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardTotalVolume
Indicates the final volume in the assay tube prior to loading onto AssayContainer. Each tube contains a standard, an InternalReference, an SDSBuffer, and a ReducingAgent and/or an AlkylatingAgent.
Default Calculation: When StandardTotalVolume is set to Automatic and IncludeStandards is True, Set StandardTotalVolume to TotalVolume.
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[50*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMix
Indicates if Standards should be mixed with StandardPremadeMasterMix that includes an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Default Calculation: When StandardPremadeMasterMix is set to Automatic, it will resolve to True if any of its downstream options is specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixReagent
The premade master mix used for CESDS experiment, containing an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Default Calculation: When StandardPremadeMasterMixReagent is set to Automatic, it will resolve to the common premade mastermix reagent in samples in .
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixDiluent
The solution used to dilute the premade master mix used to its working concentration.
Default Calculation: When StandardPremadeMasterMix is set to True and StandardPremadeMasterMixDiluent is set to Automatic, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixReagentDilutionFactor
The factor by which the premade mastermix should be diluted by in the final assay tube.
Default Calculation: When StandardPremadeMasterMix is set to True and StandardPremadeMasterMixReagentDilutionFactor is set to Automatic, it will be set as the ratio of the total volume to premade mastermix volume.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixVolume
The volume of the PremadeMasterMix required to reach its final concentration.
Default Calculation: When volume is set to Automatic, the volume is calculated by the division of TotalVolume by StandardPremadeMasterMixReagentDilutionFactor. If StandardPremadeMasterMix is set to False, StandardPremadeMasterMixVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixStorageCondition
The non-default storage condition for StandardPremadeMasterMix of this experiment after the protocol is completed. If left unset, StandardPremadeMasterMix will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardInternalReference
The solution containing the reference analyte by which Relative Migration Time is normalized. By default a 10 KDa marker is used. It is highly recommended to include an internal standard in Sample Preparation.
Default Calculation: When StandardPremadeMasterMix is set to False and InternalReference is set to Automatic, StandardInternalReference will be set as Model[Sample, StockSolution"Resuspended CESDS Internal Standard 25X"]. If StandardPremadeMasterMix is set to True, StandardInternalReference is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardInternalReferenceDilutionFactor
Marks how concentrated the internal standard is. For example, Model[Sample, StockSolution"Resuspended CESDS Internal Standard 25X"] is concentrated 25X, hence 2
µL should be added in Sample Preparation when final volume is set to 60
µL.
Default Calculation: When StandardPremadeMasterMix is set to False and StandardInternalReferenceDilutionFactor is set to Automatic, StandardInternalReferenceDilutionFactor will be set to 25. If StandardPremadeMasterMix is set to True, StandardInternalReferenceDilutionFactor is Null.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
Index Matches to: Standards
StandardInternalReferenceVolume
The volume of the internal standard added to each sample.
Default Calculation: When volume is set to Automatic, the volume is calculated by the division of TotalVolume by StandardInternalReferenceVolume. When StandardPremadeMasterMix is set to True, StandardInternalReferenceVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardConcentratedSDSBuffer
The SDS Buffer used to dilute standards. The final concentration of SDS in this assay must be equal to or greater than 0.5%.
Default Calculation: When StandardPremadeMasterMix is set to False and StandardConcentratedSDSBuffer is set to Automatic, StandardConcentratedSDSBuffer will be set as Model[Sample,"1% SDS in 100mM Tris, pH 9.5"]. If StandardPremadeMasterMix is set to True, StandardConcentratedSDSBuffer is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardConcentratedSDSBufferDilutionFactor
Marks how concentrated the StandardConcentratedSDSBuffer is. For example, a StandardConcentratedSDSBuffer that contains 1% SDS is concentrated 2X and will constitute half the StandardTotalVolume.
Default Calculation: When StandardPremadeMasterMix is set to False and StandardConcentratedSDSBufferDilutionFactor is set to Automatic, StandardConcentratedSDSBufferDilutionFactor will be set to 2. If StandardPremadeMasterMix is set to True, StandardConcentratedSDSBufferDilutionFactor is Null.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
Index Matches to: Standards
StandardDiluent
The solution used to dilute StandardConcentratedSDSBuffer to working concentration.
Default Calculation: When StandardPremadeMasterMix is set to False and Diluent is set to Automatic, Diluent will be set to Model[Sample,"Milli-Q water"]. If StandardPremadeMasterMix is set to True, StandardDiluent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardConcentratedSDSBufferVolume
The volume of StandardConcentratedSDSBufferVolume added to each sample.
Default Calculation: When volume is set to Automatic and StandardPremadeMasterMix is set to False, the volume is calculated by the division of StandardTotalVolume by StandardConcentratedSDSBufferDilutionFactor. When StandardPremadeMasterMix is set to True, StandardConcentratedSDSBufferVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardSDSBuffer
The SDS Buffer used to dilute the sample. The final concentration of SDS in this assay must be equal or greater than 0.5%.
Default Calculation: When StandardSDSBuffer is set to Automaticit is resolved to Null, so that Concentrated SDSBuffer, if set to Automatic, is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardSDSBufferVolume
The volume of SDSBuffer added to each sample.
Default Calculation: When StandardSDSBufferVolume is set to Automatic and StandardSDSBuffer is not Null, the volume is calculated by the difference between the StandardTotalVolume and the volume in the tube that includes the Standard, InternalReference, ReducingAgent, and AlkylatingAgent.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardReduction
Indicates if disulfide bridges should be chemically reduced in the Standard.
Default Calculation: When automatic, StandardReduction will set to True if any of the Reduction options are not Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standards
StandardReducingAgent
The reducing agent to be added to the Standard. for example,
β-mercaptoethanol or Dithiothreitol.
Default Calculation: When StandardPremadeMasterMix is set to False and StandardReducingAgent is set to Automatic, StandardReducingAgent will be set to Model[Sample,"2-mercaptoethanol"]. If StandardPremadeMasterMix is set to True, StandardReducingAgent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardReducingAgentTargetConcentration
The final concentration of the reducing agent in the Standard.
Default Calculation: When target concentration is set to Automatic, and no StandardReducingAgentVolume value is given, concentration is set to 650 mM. If volume is given concentration will be calculated. If StandardPremadeMasterMix is set to True, StandardReducingAgentTargetConcentration is Null
Pattern Description: Greater than 0 molar or greater than or equal to 0 VolumePercent and less than or equal to 100 VolumePercent or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | RangeP[0*VolumePercent, 100*VolumePercent]) | Automatic) | Null
Index Matches to: Standards
StandardReducingAgentVolume
The volume of the reducing agent required to reach its final concentration in the Standard.
Default Calculation: When StandardReducingAgentVolume is set to Automatic and StandardPremadeMasterMix is set to False, calculate the volume required to reach StandardReducingAgentTargetConcentration, if set. When StandardReducingAgentVolume is set to Automatic and StandardReducingAgentTargetConcentration is set to Null, StandardReducingAgentVolume is set to Null. When StandardPremadeMasterMix is set to True, StandardReducingAgentVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardAlkylation
Indicates if Alkylation should be applied to the Standard. Alkylation of free cysteines is useful in mitigating unexpected protein fragmentation and reproducibility issues.
Default Calculation: When automatic, StandardAlkylation will set to True if any of the Reduction options are not Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standards
StandardAlkylatingAgent
The alkylating agent to be added to the Standard. For example, Iodoacetamide.
Default Calculation: When StandardPremadeMasterMix is set to False and StandardAlkylatingAgent is set to Automatic, StandardAlkylatingAgent will be set to Model[Sample,"250 mM Iodoacetamide"]. If StandardPremadeMasterMix is set to True, StandardAlkylatingAgent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardAlkylatingAgentTargetConcentration
The final concentration of the alkylating agent in the Standard.
Default Calculation: When target concentration is set to Automatic, StandardPremadeMasterMix is set to False, and no StandardAlkylatingAgentVolume value is given, concentration is set to 11.5 millimolar. If volume is given concentration will be calculated. If StandardPremadeMasterMix is set to True, StandardAlkylatingAgentTargetConcentration is Null.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: Standards
StandardAlkylatingAgentVolume
The volume of the alkylating agent required to reach its final concentration in the Standard.
Default Calculation: When StandardAlkylatingAgentVolume is set to Automatic and StandardPremadeMasterMix is set to False, calculate the volume required to reach StandardAlkylatingAgentTargetConcentration, if set. When StandardAlkylatingAgentVolume is set to Automatic and StandardAlkylatingAgentTargetConcentration is set to Null, StandardAlkylatingAgentVolume is set to Null. If StandardPremadeMasterMix is set to True, StandardAlkylatingAgentVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardSedimentationSupernatantVolume
The volume of supernatant to transfer to the assay container from the Standard tubes after denaturation and centrifugation. A minimum of 50
µL are required for the analysis to proceed.
Default Calculation: When StandardPelletSedimentation is True and SedimentationSupernatantVolume is set to Automatic, 90% of the TotalVolume is drawn to avoid disturbing the pellet.
Pattern Description: Greater than or equal to 50 microliters or Null.
Programmatic Pattern: (GreaterEqualP[50*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardInternalReferenceStorageCondition
The non-default storage condition for StandardInternalReference of this experiment after the protocol is completed. If left unset, StandardInternalReferences will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardConcentratedSDSBufferStorageCondition
The non-default storage condition for StandardConcentratedSDSBuffer of this experiment after the protocol is completed. If left unset, StandardConcentratedSDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardDiluentStorageCondition
The non-default storage condition for StandardDiluent of this experiment after the protocol is completed. If left unset, StandardDiluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardSDSBufferStorageCondition
The non-default storage condition for StandardSDSBuffer of this experiment after the protocol is completed. If left unset, StandardSDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardReducingAgentStorageCondition
The non-default storage condition for StandardReducingAgent of this experiment after the protocol is completed. If left unset, StandardReducingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardAlkylatingAgentStorageCondition
The non-default storage condition for StandardAlkylatingAgent of this experiment after the protocol is completed. If left unset, StandardAlkylatingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
Standard Separation
StandardInjectionVoltageDurationProfile
Voltage profile for Injection. Supports up to 20 steps where each step is 0-200 Second, and 0-6500 volts.
Default Calculation: When StandardInjectionVoltageDurationProfile is set to Automatic and IncludeStandards is True, StandardInjectionVoltageDurationProfile is set to {{4600*Volt,20*Second}}, otherwise, Null.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: ({{RangeP[0*Volt, 6500*Volt], RangeP[0*Second, 200*Second]}..} | Automatic) | Null
Index Matches to: Standards
StandardSeparationVoltageDurationProfile
Voltage profile for Injection. Supports up to 20 steps.
Default Calculation: When StandardSeparationVoltageDurationProfile is set to Automatic and IncludeStandards is True, separation time will be set according to whether or not standards are reduced or not, and which Standard is used. If standard is IgG and not reduced, set time to 25 minutes, otherwise set to 35 minutes. For all conditions, voltage is set to 5750 Volts. When IncludeStandards is False, StandardSeparationVoltageDurationProfile is Null.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: ({{RangeP[0*Volt, 6500*Volt], RangeP[0*Minute, 180*Minute]}..} | Automatic) | Null
Index Matches to: Standards
Blanks
IncludeBlanks
Indicates if standards should be included in this experiment. Standards contain identified analytes of known size and Relative Migration Times that can be used as reference for unknown analytes.
Default Calculation: If any Blank-related options are set, then this is set True; otherwise, false.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Blanks
Indicates which Blanks to include.
Default Calculation: If IncludeStandards is True, set to CESDS Sample Buffer.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankStorageCondition
The non-default storage condition for Blanks of this experiment after the protocol is completed. If left unset, BlanksStorageCondition will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankFrequency
Indicates how many injections per Blank should be included in this experiment. Sample, Standard, and Blank injection order are resolved according to InjectionTable.
Default Calculation: When BlankFrequency is set to Automatic and IncludeBlanks is True, Set Frequency to FirstAnLast.
Pattern Description: _?(GreaterQ[#1, 0, 1] & ), FirstAndLast, First, or Last or Null.
Programmatic Pattern: ((GreaterP[0, 1] | FirstAndLast | First | Last) | Automatic) | Null
Blank Preparation
BlankVolume
Indicates the volume drawn from the blank to the assay tube. Each tube contains a Blank, an InternalReference, an SDSBuffer, and a ReducingAgent and/or an AlkylatingAgent.
Default Calculation: When BlankVolume is set to Automatic and IncludeBlanks is True, the volume is calculated to be 25% of the BlankTotalVolume. When IncludeBlanks is False, BlankVolume is Null. If InjectionTable is informed, specified volumes will be used.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
BlankTotalVolume
Indicates the final volume in the assay tube prior to loading onto AssayContainer. Each tube contains a standard, an InternalReference, an SDSBuffer, and a ReducingAgent and/or an AlkylatingAgent.
Default Calculation: When BlankTotalVolume is set to Automatic and IncludeBlanks is True, Set BlankTotalVolume to TotalVolume. When IncludeBlanks is set to False, BlankTotalVolume is Null.
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[50*Microliter, 200*Microliter] | Automatic) | Null
BlankPremadeMasterMix
Indicates if Blanks should be mixed with StandardPremadeMasterMix that includes an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Default Calculation: When BlankPremadeMasterMix is set to Automatic, it will resolve to True if any of its downstream options is specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankPremadeMasterMixReagent
The premade master mix used for CESDS experiment, containing an SDS buffer, an internal standard, and reducing and / or alkylating agents (if applicable).
Default Calculation: When BlankPremadeMasterMixReagent is set to Automatic, it will resolve to the common premade mastermix reagent in samples in .
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankPremadeMasterMixDiluent
The solution used to dilute the premade master mix used to its working concentration.
Default Calculation: When BlankPremadeMasterMix is set to True and BlankPremadeMasterMixDiluent is set to Automatic, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankPremadeMasterMixReagentDilutionFactor
The factor by which the premade mastermix should be diluted by in the final assay tube.
Default Calculation: When PremadeMasterMix is set to True and PremadeMasterMixReagentDilutionFactor is set to Automatic, it will be set as the ratio of the total volume to premade mastermix volume.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
BlankPremadeMasterMixVolume
The volume of the premade mastermix required to reach its final concentration.
Default Calculation: When volume is set to Automatic, the volume is calculated by the division of TotalVolume by BlankPremadeMasterMixReagentDilutionFactor. If BlankPremadeMasterMix is set to False, BlankPremadeMasterMixVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
BlankPremadeMasterMixStorageCondition
The non-default storage condition for BlankPremadeMasterMix of this experiment after the protocol is completed. If left unset, BlankPremadeMasterMix will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankInternalReference
The solution containing the reference analyte by which Relative Migration Time is normalized. By default a 10 KDa marker is used. It is highly recommended to include an internal standard in Sample Preparation.
Default Calculation: When BlankPremadeMasterMix is set to False and InternalReference is set to Automatic, BlankInternalReference will be set as Model[Sample, StockSolution"Resuspended CESDS Internal Standard 25X"]. If BlankPremadeMasterMix is set to True, BlankInternalReference is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankInternalReferenceDilutionFactor
Marks how concentrated the internal standard is. For example, Model[Sample, StockSolution"Resuspended CESDS Internal Standard 25X"] is concentrated 25X, hence 2
µL should be added in Sample Preparation when final volume is set to 60
µL.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankInternalReferenceDilutionFactor is set to Automatic, BlankInternalReferenceDilutionFactor will be set to 25. If BlankPremadeMasterMix is set to True, BlankInternalReferenceDilutionFactor is Null.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
BlankInternalReferenceVolume
The volume of the internal standard added to each blank.
Default Calculation: When volume is set to Automatic, the volume is calculated by the division of TotalVolume by StandardInternalReferenceVolume. When BlankPremadeMasterMix is set to True, BlankInternalReferenceVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
BlankConcentratedSDSBuffer
The SDS Buffer used to dilute blanks. The final concentration of SDS in this assay must be equal to or greater than 0.5%.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankConcentratedSDSBuffer is set to Automatic, BlankConcentratedSDSBuffer will be set as Model[Sample,"1% SDS in 100mM Tris, pH 9.5"]. If BlankPremadeMasterMix is set to True, BlankConcentratedSDSBuffer is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankConcentratedSDSBufferDilutionFactor
Marks how concentrated the BlankConcentratedSDSBuffer is. For example, a BlankConcentratedSDSBuffer that contains 1% SDS is concentrated 2X and will constitute half the BlankTotalVolume.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankConcentratedSDSBufferDilutionFactor is set to Automatic, BlankConcentratedSDSBufferDilutionFactor will be set to 2. If BlankPremadeMasterMix is set to True, BlankConcentratedSDSBufferDilutionFactor is Null.
Pattern Description: Greater than 1 or Null.
Programmatic Pattern: (GreaterP[1] | Automatic) | Null
BlankDiluent
The solution used to dilute BlankConcentratedSDSBuffer to working concentration.
Default Calculation: When BlankPremadeMasterMix is set to False and Diluent is set to Automatic, Diluent will be set to Model[Sample,"Milli-Q water"]. If BlankPremadeMasterMix is set to True, BlankDiluent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankConcentratedSDSBufferVolume
The volume of BlankConcentratedSDSBufferVolume added to each sample.
Default Calculation: When volume is set to Automatic and BlankPremadeMasterMix is set to False, the volume is calculated by the division of BlankTotalVolume by BlankConcentratedSDSBufferDilutionFactor. When BlankPremadeMasterMix is set to True, BlankConcentratedSDSBufferVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
BlankSDSBuffer
The SDS Buffer used to dilute the sample. The final concentration of SDS in this assay must be equal or greater than 0.5%.
Default Calculation: When BlankSDSBuffer is set to Automaticit is resolved to Null, so that Concentrated SDSBuffer, if set to Automatic, is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankSDSBufferVolume
The volume of SDSBuffer added to each sample.
Default Calculation: When BlankSDSBufferVolume is set to Automatic and BlankSDSBuffer is not Null, the volume is calculated by the difference between the BlankTotalVolume and the volume in the tube that includes the Blank, InternalReference, ReducingAgent, and AlkylatingAgent.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
BlankReduction
Indicates if disulfide bridges should be chemically reduced in the Blank.
Default Calculation: When automatic, StandardReduction will set to True if any of the Reduction options are not Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankReducingAgent
The reducing agent to be added to the Blank. for example,
β-mercaptoethanol or Dithiothreitol.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankReducingAgent is set to Automatic, BlankReducingAgent will be set to Model[Sample,"2-mercaptoethanol"]. If BlankPremadeMasterMix is set to True, BlankReducingAgent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankReducingAgentTargetConcentration
The final concentration of the reducing agent in the Blank.
Default Calculation: When target concentration is set to Automatic, and no BlankReducingAgentVolume value is given, concentration is set to 650 mM. If volume is given concentration will be calculated. If BlankPremadeMasterMix is set to True, BlankReducingAgentTargetConcentration is Null
Pattern Description: Greater than 0 molar or greater than or equal to 0 VolumePercent and less than or equal to 100 VolumePercent or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | RangeP[0*VolumePercent, 100*VolumePercent]) | Automatic) | Null
BlankReducingAgentVolume
The volume of the reducing agent required to reach its final concentration in the Blank.
Default Calculation: When BlankReducingAgentVolume is set to Automatic and BlankPremadeMasterMix is set to False, calculate the volume required to reach BlankReducingAgentTargetConcentration, if set. When BlankReducingAgentVolume is set to Automatic and BlankReducingAgentTargetConcentration is set to Null, BlankReducingAgentVolume is set to Null. When BlankPremadeMasterMix is set to True, BlankReducingAgentVolume is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
BlankAlkylation
Indicates if Alkylation should be applied to the Blank. Alkylation of free cysteines is useful in mitigating unexpected protein fragmentation and reproducibility issues.
Default Calculation: When automatic, BlankAlkylation will set to True if any of the Reduction options are not Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankAlkylatingAgent
The alkylating agent to be added to the Blank. For example, Iodoacetamide.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankAlkylatingAgent is set to Automatic, BlankAlkylatingAgent will be set to Model[Sample,"250 mM Iodoacetamide"]. If BlankPremadeMasterMix is set to True, BlankAlkylatingAgent is Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankAlkylatingAgentTargetConcentration
The final concentration of the alkylating agent in the Blank.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankAlkylatingAgent is set to Automatic, BlankAlkylatingAgent will be set to Model[Sample,"250 mM Iodoacetamide"]. If BlankPremadeMasterMix is set to True, BlankAlkylatingAgent is Null.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
BlankAlkylatingAgentVolume
The volume of the alkylating agent required to reach its final concentration in the Blank.
Default Calculation: When target concentration is set to Automatic, BlankPremadeMasterMix is set to False, and no BlankAlkylatingAgentVolume value is given, concentration is set to 11.5 millimolar. If volume is given concentration will be calculated. If BlankPremadeMasterMix is set to True, BlankAlkylatingAgentTargetConcentration is Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
BlankSedimentationSupernatantVolume
The volume of supernatant to transfer to the assay container from the Blank tubes after denaturation and centrifugation. A minimum of 50
µL are required for the analysis to proceed.
Default Calculation: When BlankPelletSedimentation is True and SedimentationSupernatantVolume is set to Automatic, 90% of the TotalVolume is drawn to avoid disturbing the pellet.
Pattern Description: Greater than or equal to 50 microliters or Null.
Programmatic Pattern: (GreaterEqualP[50*Microliter] | Automatic) | Null
BlankInternalReferenceStorageCondition
The non-default storage condition for BlankInternalReference of this experiment after the protocol is completed. If left unset, BlankInternalReferences will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankConcentratedSDSBufferStorageCondition
The non-default storage condition for BlankConcentratedSDSBuffer of this experiment after the protocol is completed. If left unset, BlankConcentratedSDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankDiluentStorageCondition
The non-default storage condition for BlankDiluent of this experiment after the protocol is completed. If left unset, BlankDiluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankSDSBufferStorageCondition
The non-default storage condition for BlankSDSBuffer of this experiment after the protocol is completed. If left unset, BlankSDSBuffer will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankReducingAgentStorageCondition
The non-default storage condition for BlankReducingAgent of this experiment after the protocol is completed. If left unset, BlankReducingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankAlkylatingAgentStorageCondition
The non-default storage condition for BlankAlkylatingAgent of this experiment after the protocol is completed. If left unset, BlankAlkylatingAgent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Blank Separation
BlankInjectionVoltageDurationProfile
Voltage profile for Injection. Supports up to 20 steps where each step is 0-200 Second, and 0-6500 volts.
Default Value: {{4600 volts, 20 seconds}}
Default Calculation: When BlankInjectionVoltageDurationProfile is set to Automatic and IncludeBlanks is True, BlankInjectionVoltageDurationProfile is set to {{4600*Volt,20*Second}}, otherwise, it is set to Null.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: {{RangeP[0*Volt, 6500*Volt], RangeP[0*Second, 200*Second]}..} | Null
BlankSeparationVoltageDurationProfile
Voltage profile for Injection. Supports up to 20 steps.
Default Calculation: When BlankSeparationVoltageDurationProfile is set to Automatic, separation time will be set according to whether or not standards are reduced or not, and which Blank is used. If standard is IgG and not reduced, set time to 25 minutes, otherwise set to 35 minutes. For all conditions, voltage is set to 5750 Volts. If IncludeBlanks is False, BlankSeparationVoltageDurationProfile is set to Null.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: ({{RangeP[0*Volt, 6500*Volt], RangeP[0*Minute, 180*Minute]}..} | Automatic) | Null
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and when the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise set to Model[Container, Vessel, "2mL Tube"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[Model[Container]] | Automatic) | Null
Index Matches to: experiment samples
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
Index Matches to: experiment samples
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples