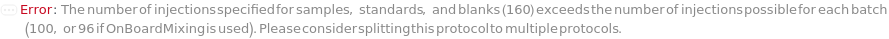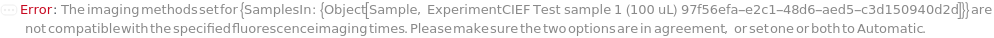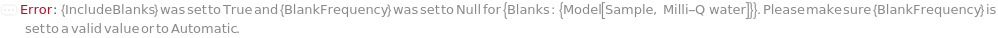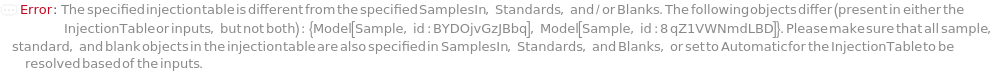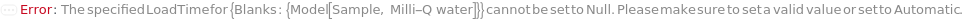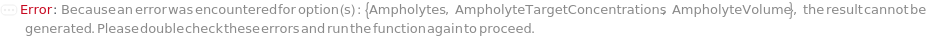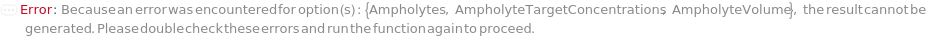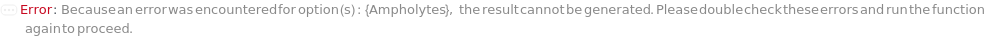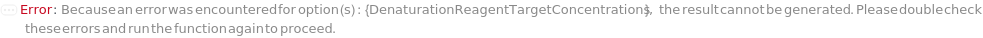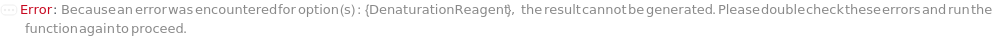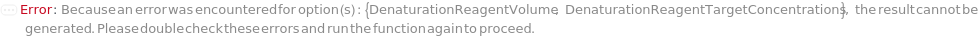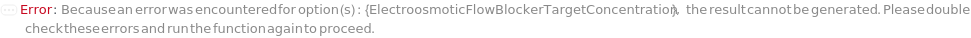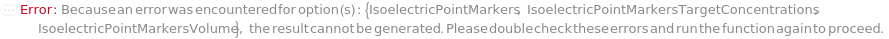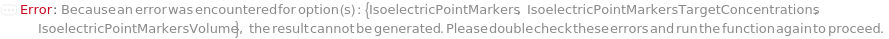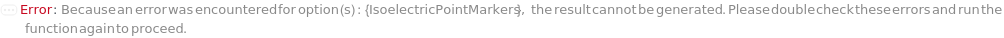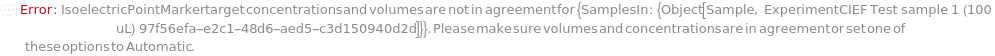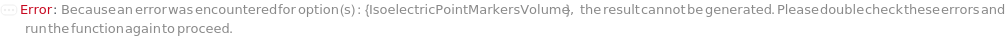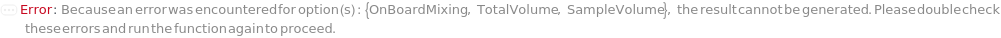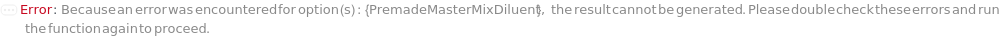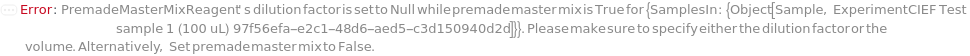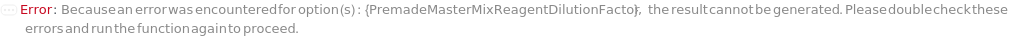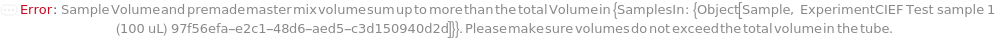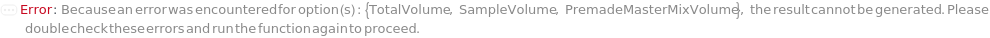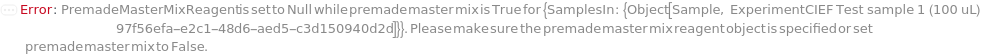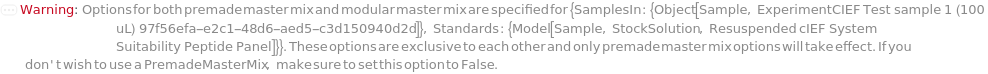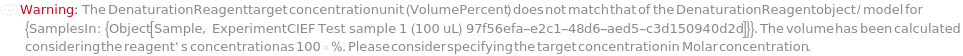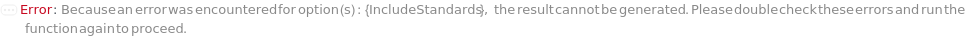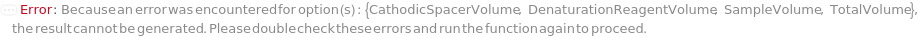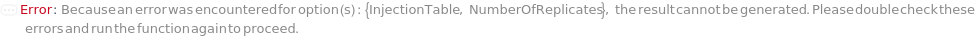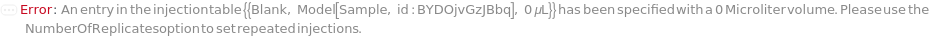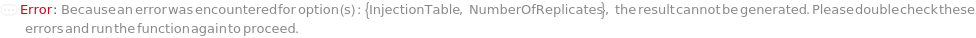General
Instrument
The capillary electrophoresis instrument that will be used by the protocol. The instrument accepts a capillary cartridge loaded with electrolytes and sequentially analyzes samples by separating proteins according to their isoelectric point in a pH gradient.
Default Value: Model[Instrument, ProteinCapillaryElectrophoresis, Maurice]
Pattern Description: An object of type or subtype Model[Instrument, ProteinCapillaryElectrophoresis] or Object[Instrument, ProteinCapillaryElectrophoresis]
Programmatic Pattern: ObjectP[{Model[Instrument, ProteinCapillaryElectrophoresis], Object[Instrument, ProteinCapillaryElectrophoresis]}]
Cartridge
The capillary electrophoresis cartridge loaded on the instrument for Capillary IsoElectric Focusing (cIEF) experiments. The cartridge holds a single capillary and electrolyte buffers (sources of hydronium and hydroxyl ions). The cIEF cartridge can run 100 injections in up to 20 batches.
Default Value: Model[Container, ProteinCapillaryElectrophoresisCartridge, cIEF]
Pattern Description: An object of type or subtype Model[Container, ProteinCapillaryElectrophoresisCartridge] or Object[Container, ProteinCapillaryElectrophoresisCartridge] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Container, ProteinCapillaryElectrophoresisCartridge], Object[Container, ProteinCapillaryElectrophoresisCartridge]}] | _String
SampleTemperature
The sample tray temperature at which samples are maintained while awaiting injection.
Default Value: 10 degrees Celsius
Pattern Description: Ambient, 4 degrees Celsius, 10 degrees Celsius, 15 degrees Celsius, or 25 degrees Celsius.
Programmatic Pattern: Ambient | 4*Celsius | 10*Celsius | 15*Celsius | 25*Celsius
InjectionTable
The order of sample, Standard, and Blank sample loading into the Instrument during measurement.
Default Calculation: Determined to the order of input samples articulated. Standard and Blank samples are inserted based on the determination of StandardFrequency and BlankFrequency. For example, StandardFrequency -> FirstAndLast and BlankFrequency -> Null result in Standard samples injected first, then samples, and then the Standard sample set again at the end.
Pattern Description: List of one or more {Type, Sample, Volume} entries.
Programmatic Pattern: {{Sample | Standard | Blank, ObjectP[{Model[Sample], Object[Sample]}] | _String, RangeP[0*Microliter, 200*Microliter] | Automatic}..} | Automatic
NumberOfReplicates
The number of times each sample will be injected. For example, when NumberOfReplicates is set to 2, each sample will be run twice consecutively. By default, this option means technical replicates that are injected from the same position on the assay plate.
Pattern Description: Greater than or equal to 1 in increments of 1 or Null.
Programmatic Pattern: (GreaterEqualP[1, 1] | Automatic) | Null
Instrument Preparation
Anolyte
The anode electrolyte solution loaded on the cartridge that is the source of hydronium ion for the capillary in cIEF experiments. Two milliliters of anolyte solution will be loaded onto the cartridge for every batch of up to 100 injections.
Default Value: Model[Sample, 0.08M Phosphoric Acid in 0.1% Methyl Cellulose]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
Catholyte
The electrolyte solution loaded on the cartridge that is the source of hydroxyl ions for the capillary in cIEF experiments. Two milliliters of catholyte solution will be loaded onto the cartridge for every batch of up to 100 injections.
Default Value: Model[Sample, 0.1M Sodium Hydroxide in 0.1% Methyl Cellulose]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
ElectroosmoticConditioningBuffer
The ElectroosmoticConditioningBuffer solution is used to wash the capillary between injections to decrease electroosmotic flow. Two milliliters of 0.5% Methyl Cellulose solution will be loaded on the instrument for every batch of up to 100 injections.
Default Value: Model[Sample, 0.5% Methyl Cellulose]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
FluorescenceCalibrationStandard
The FluorescenceCalibrationStandard solution is used to set the baseline for NativeFluorescence detection. Five hundred microliters of a commercial standard are loaded on the instrument for every batch of up to 100 injections.
Default Value: Model[Sample, cIEF Fluorescence Calibration Standard]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
WashSolution
The WashSolution is used to rinse the capillary after use. WashSolution is loaded on the instrument in two separate with 2 mL each. One vial is used to wash the capillary and the other to wash the capillary tip.
Default Value: Model[Sample, id:8qZ1VWNmdLBD]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
OnBoardMixing
Indicates whether samples should be mixed with the master mix right before they are loaded to the capillary or during sample preparation before the assay plate is loaded to the instrument. OnBoardMixing should be used for sensitive samples. Only a single master mix composition is allowed when using OnBoardMixing. When using OnBoardMixing, Sample tubes should contain samples in 25 microliters. Before injecting each sample, the instrument will add 100 microliters of the master mix and mix.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
Post Experiment
CartridgeStorageCondition
The non-default storage condition for the Cartridge after the protocol is completed. If left unset, Cartridge will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
AnolyteStorageCondition
The non-default storage condition for the Anolyte solution after the protocol is completed. If left unset, Cartridge will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
CatholyteStorageCondition
The non-default storage condition for the Catholyte solution after the protocol is completed. If left unset, Cartridge will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
ElectroosmoticConditioningBufferStorageCondition
The non-default storage condition for the ElectroosmoticConditioningBuffer after the protocol is completed. If left unset, Cartridge will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
FluorescenceCalibrationStandardStorageCondition
The non-default storage condition for the FluorescenceCalibrationStandard after the protocol is completed. If left unset, Cartridge will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
WashSolutionStorageCondition
The non-default storage condition for the WashSolution after the protocol is completed. If left unset, Cartridge will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
AmpholytesStorageCondition
The non-default storage condition for ampholyte stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Default Calculation: When Automatic, resolve according to the resolved ampholytes.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null entries.
Programmatic Pattern: (((SampleStorageTypeP | Disposal | Null) | {(SampleStorageTypeP | Disposal | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
IsoelectricPointMarkersStorageCondition
The non-default storage condition for IsoelectricPointMarker stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Default Calculation: When Automatic, resolve according to the resolved isoelectricPointMarkers.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null entries.
Programmatic Pattern: (((SampleStorageTypeP | Disposal | Null) | {(SampleStorageTypeP | Disposal | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
DenaturationReagentStorageCondition
The non-default storage condition for DenaturationReagent stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
AnodicSpacerStorageCondition
The non-default storage condition for AnodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
CathodicSpacerStorageCondition
The non-default storage condition for CathodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
ElectroosmoticFlowBlockerStorageCondition
The non-default storage condition for CathodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
DiluentStorageCondition
The non-default storage condition for Diluent of this experiment after the protocol is completed. If left unset, Diluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
StandardStorageCondition
The non-default storage condition for Standards of this experiment after the protocol is completed. If left unset, StandardStorageCondition will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardAmpholytesStorageCondition
The non-default storage condition for ampholyte stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Default Calculation: When Automatic and IncludeStandards is True, resolves to Null according to the resolved ampholytes.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null entries or Null.
Programmatic Pattern: (((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal | Null)..}) | Automatic) | Null
Index Matches to: Standards
StandardIsoelectricPointMarkersStorageCondition
The non-default storage condition for IsoelectricPointMarker stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Default Calculation: When Automatic and IncludeStandards is True, resolves to Null according to the resolved isoelectricPointMarkers.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null entries or Null.
Programmatic Pattern: (((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal | Null)..}) | Automatic) | Null
Index Matches to: Standards
StandardDenaturationReagentStorageCondition
The non-default storage condition for DenaturationReagent stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardAnodicSpacerStorageCondition
The non-default storage condition for AnodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardCathodicSpacerStorageCondition
The non-default storage condition for CathodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardElectroosmoticFlowBlockerStorageCondition
The non-default storage condition for CathodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
StandardDiluentStorageCondition
The non-default storage condition for Diluent of this experiment after the protocol is completed. If left unset, Diluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: Standards
BlankStorageCondition
The non-default storage condition for Blanks of this experiment after the protocol is completed. If left unset, BlanksStorageCondition will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankAmpholytesStorageCondition
The non-default storage condition for ampholyte stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Default Calculation: When Automatic, resolve according to the resolved ampholytes.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null entries or Null.
Programmatic Pattern: (((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal | Null)..}) | Automatic) | Null
BlankIsoelectricPointMarkersStorageCondition
The non-default storage condition for IsoelectricPointMarker stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Default Calculation: When Automatic, resolve according to the resolved isoelectricPointMarkers.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or list of one or more {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting}, Disposal, or Null entries or Null.
Programmatic Pattern: (((SampleStorageTypeP | Disposal) | {(SampleStorageTypeP | Disposal | Null)..}) | Automatic) | Null
BlankDenaturationReagentStorageCondition
The non-default storage condition for DenaturationReagent stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankAnodicSpacerStorageCondition
The non-default storage condition for AnodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankCathodicSpacerStorageCondition
The non-default storage condition for CathodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankElectroosmoticFlowBlockerStorageCondition
The non-default storage condition for CathodicSpacer stocks of this experiment after the protocol is completed. If left unset, stocks will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
BlankDiluentStorageCondition
The non-default storage condition for Diluent of this experiment after the protocol is completed. If left unset, Diluent will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples
Sample Preparation
SampleVolume
Indicates the volume drawn from the sample to the assay tube. Each tube contains a Sample, DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When SampleVolume is set to Automatic and OnBoardMixing is False, the volume is calculated based off of the composition of the sample to reach 0.2 mg / ml protein in the sample. If composition is not available, volume will be set to 10% of the TotalVolume. When SampleVolume is set to Automatic and OnBoardMixing is True, SampleVolume is resolves to 25 microliters.
Pattern Description: Greater than 0 microliters.
Programmatic Pattern: GreaterP[0*Microliter] | Automatic
Index Matches to: experiment samples
TotalVolume
Indicates the final volume in the assay tube prior to loading onto the capillary. Each tube contains a Sample, DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When TotalVolume is set to Automatic and OnBoardMixing is True, Total Volume is resolved to 125 microliters. OnBoardMixing is False, Total volume is defaulted to 100 microliters.
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 200 microliters.
Programmatic Pattern: RangeP[50*Microliter, 200*Microliter] | Automatic
Index Matches to: experiment samples
PremadeMasterMix
Indicates if a premade master mix should be used or, alternatively, the master mix should be made as part of this protocol. The master mix contains the reagents required for cIEF experiments, i.e., DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When PremadeMasterMix is set to Automatic, it will resolve to True if any of its downstream options is specified.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
PremadeMasterMixReagent
The premade master mix used for cIEF experiment, containing DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When PremadeMasterMix is True and PremadeMasterMixReagent is set to Automatic, it will resolve to Model[Sample, StockSolution, "2X Wide-Range cIEF Premade Master Mix"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
PremadeMasterMixDiluent
The solution used to dilute the premade master mix used to its working concentration.
Default Calculation: When PremadeMasterMix is set to True and PremadeMasterMixDiluent is set to Automatic, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
PremadeMasterMixReagentDilutionFactor
The factor by which the premade master mix should be diluted by in the final assay tube.
Default Calculation: When PremadeMasterMix is set to True and PremadeMasterMixReagentDilutionFactor is set to Automatic, it will be set as the ratio of the total volume to premade master mix volume.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
Index Matches to: experiment samples
PremadeMasterMixVolume
The volume of the premade master mix required to reach its final concentration.
Default Calculation: When PremadeMasterMix is set to True and PremadeMasterMixVolume is set to Automatic, the volume is calculated by the division of TotalVolume by PremadeMasterMixReagentDilutionFactor. If PremadeMasterMix is set to False, PremadeMasterMixVolume is Null.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
Diluent
The solution used to top volume in assay tube to total volume and dilute components to working concentration.
Default Calculation: When PremadeMasterMix is set to False and Diluent is set to Automatic, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
Ampholytes
A solution composed of a mixture of amphoteric molecules, that case act both as an acid and a base, in the master mix that form the pH gradient. In the presence of an anolyte and a catholyte, and once a voltage is applied across the capillary, ampholytes for a pH gradient at a range that depend on the amopholyte composition. Proteins then resolve according to their isoelectric point within this gradient. When specifying values for each of SamplesIn, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When Ampholytes is set to Automatic and PremadeMasterMix is False, Model[Sample,"Pharmalyte pH 3-10"] will be set as the ampholyte in this sample.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or {Automatic, Null} entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Sample], Object[Sample]}] | _String) | {((ObjectP[{Model[Sample], Object[Sample]}] | _String) | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: experiment samples
AmpholyteTargetConcentrations
The concentration (Vol/Vol) of amphoteric molecules in the master mix. When specifying values for each of SamplesIn, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When PremadeMasterMix is set to False and AmpholyteTargetConcentrations is set to Automatic, AmpholyteTargetConcentrations will be set to 4% if one ampholyte is given, and 2% each, if more than one ampholyte is set and no AmpholyteVolume values are given. If volume is given concentration will be calculated considering Ampholyte concentrations as 100%.
Pattern Description: Greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or list of one or more {Automatic, Null} or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent entries or Null.
Programmatic Pattern: ((RangeP[0.001*VolumePercent, 100*VolumePercent] | {(RangeP[0.001*VolumePercent, 100*VolumePercent] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: experiment samples
AmpholyteVolume
The volume of amphoteric molecule stocks to add to the master mix. When specifying values for each of SamplesIn, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When PremadeMasterMix is set to False and AmpholyteVolume is set to Automatic, AmpholyteVolume is set to the volume needed to reach the given AmpholyteTargetConcentrations based in the TotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or list of one or more {Automatic, Null} or greater than or equal to 0.1 microliters and less than or equal to 200 microliters entries or Null.
Programmatic Pattern: ((RangeP[0.1*Microliter, 200*Microliter] | {(RangeP[0.1*Microliter, 200*Microliter] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: experiment samples
IsoelectricPointMarkers
Reference analytes, usually peptides with known isoelectric points, that are included in the master mix and are used to convert position in the capillary to the pI it represents. The mastermix for each sample should have two isoelectric point markers. A linear fit is then used to directly interpret the pI for each position between the two markers. When specifying values for each of SamplesIn, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When IsoelectricPointMarkers is set to Automatic and PremadeMasterMix is False, {Model[Sample,StockSolution, "Resuspended cIEF pI Marker - 4.05"], Model[Sample,StockSolution, "Resuspended cIEF pI Marker - 9.99"]} will be set as the IsoelectricPointMarkers in this sample.
Pattern Description: List of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample entries or list of one or more {Automatic, Null} entries or Null.
Programmatic Pattern: (({(ObjectP[{Model[Sample], Object[Sample]}] | _String)..} | {(Automatic | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
IsoelectricPointMarkersTargetConcentrations
The final concentration (Vol/Vol) of pI markers in the assay tube. When specifying values for each of SamplesIn, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When IsoelectricPointMarkers is set to Automatic and PremadeMasterMix is False, pI markers target concentration will be set to 1%.
Pattern Description: Greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or list of one or more {Automatic, Null} or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent entries or Null.
Programmatic Pattern: ((RangeP[0.001*VolumePercent, 100*VolumePercent] | {(RangeP[0.001*VolumePercent, 100*VolumePercent] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: experiment samples
IsoelectricPointMarkersVolume
The volume of pI marker stocks to add to the master mix. When specifying values for each of SamplesIn, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When IsoelectricPointMarkersVolume is set to Automatic and PremadeMasterMix is False, it is set to the volume required to reach IsoelectricPointMarkersTargetConcentrations in TotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or list of one or more {Automatic, Null} or greater than or equal to 0.1 microliters and less than or equal to 200 microliters entries or Null.
Programmatic Pattern: ((RangeP[0.1*Microliter, 200*Microliter] | {(RangeP[0.1*Microliter, 200*Microliter] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: experiment samples
ElectroosmoticFlowBlocker
Electroosmotic flow blocker solution, usually methyl cellulose, to include in the master mix. Electroosmotic flow referes to the motion of liquids in a capillary as a result of applied voltage across it. Electroosmotic flow is most significant when channel diameters are very small. In capillary isoelectric focusing, the charge of the capillary wall should be masked to minimize the electroosmotic flow.
Default Calculation: When ElectroosmoticFlowBlocker is set to Automatic and PremadeMasterMix is False, ElectroosmoticFlowBlocker will be set to Model[Sample,"1% Methyl Cellulose"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
ElectroosmoticFlowBlockerTargetConcentrations
The concentration of ElectroosmoticFlowBlocker in the master mix.
Default Calculation: When ElectroosmoticFlowBlockerTargetConcentrations is set to Automatic and PremadeMasterMix is False, it is set to 0.35% if no ElectroosmoticFlowBlockerVolume is set.
Pattern Description: Greater than or equal to 0.001 MassPercent and less than or equal to 100 MassPercent or Null.
Programmatic Pattern: (RangeP[0.001*MassPercent, 100*MassPercent] | Automatic) | Null
Index Matches to: experiment samples
ElectroosmoticFlowBlockerVolume
The volume of ElectroosmoticBlocker stock to add to the master mix.
Default Calculation: When ElectroosmoticFlowBlockerVolume is set to Automatic and PremadeMasterMix is False, it is calculated based on TotalVolume to reach the ElectroosmoticBlockerTargetConcentrations.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
Denature
Indicates if a DenaturationReagent should be added to the master mix.
Default Calculation: When Denature is set to Automatic and PremadeMasterMix is False, Denature will be set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
DenaturationReagent
The denaturing agent, e.g., Urea or SimpleSol, to be added to the master mix to prevent protein precipitation.
Default Calculation: When DenaturationReagent is set to Automatic, PremadeMasterMix is False, and Denature is True, DenaturationReagent will be set to Model[Sample,StockSolution,"8M Urea"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
DenaturationReagentTargetConcentration
The final concentration of the denaturing agent in the master mix.
Default Calculation: When DenaturationReagentTargetConcentration is set to Automatic, PremadeMasterMix is False, and Denature is True, it is set to 4M if no DenaturationReagentVolume value is given. If DenaturationReagentVolume is set and the concentration of DenaturationReagent is known, DenaturationReagentTargetConcentration is calculated.
Pattern Description: Greater than 0 molar or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | RangeP[0.001*VolumePercent, 100*VolumePercent]) | Automatic) | Null
Index Matches to: experiment samples
DenaturationReagentVolume
The volume of the denaturing agent required to reach its final concentration in the master mix.
Default Calculation: When DenaturationReagentVolume is set to Automatic, PremadeMasterMix is False, and Denature is True, it is set to the volume required to reach DenaturationReagentTargetConcentration.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
IncludeAnodicSpacer
Indicates if an anodic spacer should be added to the master mix. Both acidic and alkaline carrier ampholytes may be lost in the electrolyte reservoirs due to diffusion and isotachophoresis, decreasing the resolution and detection of proteins at the extremes of the pH gradient. To reduce the loss of ampholytes, Spacers (ampholytes with very low or very high pIs) can be added to buffer the loss of analytes of interest. Traditionally, Iminodiacetic acid (pI 2.2) and Arginine (pI 10.7) are added as Spacers.
Default Calculation: When AnodicSpacer is set to Automatic and PremadeMasterMix is False, Spacers will be set to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
AnodicSpacer
Acidic ampholyte to include in the master mix. When analyzing a protein with very acidic pI (e.g. Pepsin), one might choose to add an AnodicSpacer such as Arginine (pI 10.7), to prevent loss of signal to the anolyte reservoir.
Default Calculation: When AnodicSpacer is set to Automatic, PremadeMasterMix is False and IncludeAnodicSpacer is set to True, AnodicSpacer will be set to Model[Sample,StockSolution, "200mM Iminodiacetic acid"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
AnodicSpacerTargetConcentration
The final concentration of AnodicSpacer in the master mix.
Default Calculation: When AnodicSpacerTargetConcentration is set to Automatic, PremadeMasterMix is False and IncludeAnodicSpacer is set to True, it is set to 1 mM when no AnodicSpacerMarkersVolume is set. When AnodicSpacer concentration is known and AnodicSpacerMarkersVolume is set, Automatic calculates the final concentration based on AnodicSpacerTargetConcentrations and TotalVolume.
Pattern Description: Greater than 0 millimolar or Null.
Programmatic Pattern: (GreaterP[0*Millimolar] | Automatic) | Null
Index Matches to: experiment samples
AnodicSpacerVolume
The volume of AnodicSpacer stock to add to the master mix.
Default Calculation: When AnodicSpacerVolume is set to Automatic, PremadeMasterMix is False and IncludeAnodicSpacer is set to True, it is set to the volume required to reach AnodicSpacerTargetConcentrations in TotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
IncludeCathodicSpacer
Indicates if a cathodic spacer should be added to the master mix. Both acidic and alkaline carrier ampholytes may be lost in the electrolyte reservoirs due to diffusion and isotachophoresis, decreasing the resolution and detection of proteins at the extremes of the pH gradient. To reduce the loss of ampholytes, Spacers (ampholytes with very low or very high pIs) can be added to buffer the loss of analytes of interest. Traditionally, Iminodiacetic acid (pI 2.2) and Arginine (pI 10.7) are added as Spacers.
Default Calculation: When AnodicSpacer is set to Automatic and PremadeMasterMix is False, Spacers will be set to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
CathodicSpacer
Basic ampholyte spacer to include in the master mix. When analyzing a protein with very basic pI (e.g. Cytochrome C), one might choose to add a CathodicSpacer such as Arginine (pI 10.7), to prevent loss of signal to the catholyte reservoir.
Default Calculation: When CathodicSpacer is set to Automatic, PremadeMasterMix is False and IncludeCathodicSpacer is set to True, CathodicSpacer will be set to Model[Sample,StockSolution, "500mM Arginine"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CathodicSpacerTargetConcentration
The concentration of Cathodic spacer in the master mix.
Default Calculation: When CathodicSpacerTargetConcentration is set to Automatic, PremadeMasterMix is False and IncludeCathodicSpacer is set to True, it is set to 10 mM when no CathodicSpacerMarkersVolume is set. When CathodicSpacer concentration is known and CathodicSpacerMarkersVolume is set, Automatic calculates the final concentration based on CathodicSpacerTargetConcentrations and TotalVolume.
Pattern Description: Greater than 0 millimolar or Null.
Programmatic Pattern: (GreaterP[0*Millimolar] | Automatic) | Null
Index Matches to: experiment samples
CathodicSpacerVolume
The volume of Cathodic spacer stocks to add to the master mix.
Default Calculation: When CathodicSpacerVolume is set to Automatic, PremadeMasterMix is False and IncludeCathodicSpacer is set to True, it is set to the volume required to reach AnodicSpacerTargetConcentrations in TotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
Sample Separation
LoadTime
Time to load samples into the capillary by vacuum. The default 55 Second time is enough to load approximately 8
µl. This value should be increased only if very viscous samples are used.
Default Value: 55 seconds
Pattern Description: Greater than or equal to 1 second and less than or equal to 300 seconds.
Programmatic Pattern: RangeP[1*Second, 300*Second]
Index Matches to: experiment samples
VoltageDurationProfile
Series of voltages and durations to apply onto the capillary for separation. Supports up to 20 steps where each step is 0-600 minutes, and 0-5000 volts.
Default Value: {{1500 volts, 1 minute}, {3000 volts, 10 minutes}}
Pattern Description: List of one or more {Voltage, Time} entries.
Programmatic Pattern: {{RangeP[0.1*Volt, 5000*Volt], RangeP[0.01*Minute, 600*Minute]}..}
Index Matches to: experiment samples
Sample Detection
ImagingMethods
Whole capillary imaging by Either UVAbsorbance (280 nm) alone or both UVAbsorbance and Native Fluorescence (Ex 280 nm, Em 320-450nm).
Default Calculation: When Automatic, AbsorbanceAndFluorescnece will be set.
Pattern Description: Absorbance or AbsorbanceAndFluorescence.
Programmatic Pattern: (Absorbance | AbsorbanceAndFluorescence) | Automatic
Index Matches to: experiment samples
NativeFluorescenceExposureTime
Exposure duration for NativeFluorescence detection.
Default Calculation: When Automatic, resolves according to ImagingMethods. when ImagingMethods is set to AbsorbanceAndFluorescence, NativeFluorescenceExposureTime will be set to {3*Second,5*Second,10*Second,20*Second}.
Pattern Description: Greater than or equal to 1 second and less than or equal to 200 seconds or list of one or more greater than or equal to 1 second and less than or equal to 200 seconds entries or Null.
Programmatic Pattern: ((RangeP[1*Second, 200*Second] | {RangeP[1*Second, 200*Second]..}) | Automatic) | Null
Index Matches to: experiment samples
UVDetectionWavelength
The wavelength used for signal detection by Absorbance.
Default Value: 280 nanometers
Pattern Description: An expression that matches the pattern: 280 Nanometer.
Programmatic Pattern: 280*Nanometer
NativeFluorescenceDetectionExcitationWavelengths
The excitation wavelength used for Fluorescence detection.
Default Value: 280 nanometers
Pattern Description: An expression that matches the pattern: 280 Nanometer.
Programmatic Pattern: 280*Nanometer
NativeFluorescenceDetectionEmissionWavelengths
The emission wavelength range used for Fluorescence detection.
Default Value: {320 nanometers, 405 nanometers}
Pattern Description: An expression that matches the pattern: {320 Nanometer, 405 Nanometer}
Programmatic Pattern: {320*Nanometer, 405*Nanometer}
Standards
IncludeStandards
Indicates if standards should be included in this experiment. Standards are used to both ensure reproducibility within and between Experiments and as a reference to interpolate the isoelectric point of unknown analytes in samples.
Default Calculation: When IncludeBlanks is set to Automatic and any of its related options is specified, it is set to True. Otherwise, it is set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Standards
Indicates what analyte(s) of known isoelectric points to include as part of this experiments. Standards are treated as independent samples and serve as positive controls or references for unknown samples.
Default Calculation: When Standards is set to Automatic and IncludeStandards is True, it is set to Maurice cIEF System Suitability Peptide Panel.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardFrequency
Indicates how many injections per standard should be included in this experiment. Sample, Standard, and Blank injection order are resolved according to InjectionTable.
Default Calculation: When StandardFrequency is set to Automatic and IncludeStandards is True, it will default to FirstAndLast.
Pattern Description: _?(GreaterQ[#1, 0, 1] & ), FirstAndLast, First, or Last or Null.
Programmatic Pattern: ((GreaterP[0, 1] | FirstAndLast | First | Last) | Automatic) | Null
Index Matches to: Standards
Standard Preparation
StandardVolume
Indicates the volume drawn from the standard to the assay tube. Each tube contains a Sample, DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When StandardVolume is set to Automatic and OnBoardMixing is False, the volume is calculated based off of the composition of the sample to reach 0.2 mg / ml protein in the sample. If composition is not available, volume will be set to 10% of the TotalVolume. When StandardVolume is set to Automatic and OnBoardMixing is True, StandardVolume is resolves to 25 microliters.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardTotalVolume
Indicates the final volume in the assay tube, including standard and master mix.
Default Calculation: When StandardTotalVolume is set to Automatic and IncludeStandards is True, it will resolve to the most common total volume in SamplesIn.
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[50*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMix
Indicates if a premade should be used or, alternatively, the master should be made as part of this protocol. The contains the reagents required for cIEF experiments, i.e., DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When StandardPremadeMasterMix is set to Automatic and IncludeStandards is True, it will resolve to True if any of its downstream options is specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixReagent
The premade master mix used for cIEF experiment, containing DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When StandardPremadeMasterMix is True and StandardPremadeMasterMixReagent is set to Automatic, it will resolve to Model[Sample, StockSolution, "2X Wide-Range cIEF Premade Master Mix"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixDiluent
The solution used to dilute the premade master mix used to its working concentration.
Default Calculation: When StandardPremadeMasterMixDiluent is set to Automatic, StandardPremadeMasterMix is set to True, and IncludeStandards is True, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixReagentDilutionFactor
The factor by which the premade master mix should be diluted by in the final assay tube.
Default Calculation: When StandardPremadeMasterMixReagentDilutionFactor is set to Automatic, StandardPremadeMasterMix is set to True, and IncludeStandards is True, it will be set as the ratio of the total volume to premade master mix volume.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
Index Matches to: Standards
StandardPremadeMasterMixVolume
The volume of the premade master mix required to reach its final concentration.
Default Calculation: When StandardPremadeMasterMixVolume is set to Automatic, StandardPremadeMasterMix is set to True, and IncludeStandards is True, the volume is calculated by the division of TotalVolume by StandardPremadeMasterMixReagentDilutionFactor. If StandardPremadeMasterMix is set to False, StandardPremadeMasterMixVolume is Null.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardDiluent
The solution used to top volume in assay tube to total volume and dilute components to working concentration.
Default Calculation: When StandardPremadeMasterMix is set to False, StandardDiluent is set to Automatic, and IncludeStandards is True, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardAmpholytes
A solution composed of a mixture of amphoteric molecules, that case act both as an acid and a base, in the master mix that form the pH gradient. In the presence of an anolyte and a catholyte, and once a voltage is applied across the capillary, ampholytes for a pH gradient at a range that depend on the amopholyte composition. Proteins then resolve according to their isoelectric point within this gradient. When specifying values for each of Standards, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When StandardAmpholytes is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, Model[Sample,"Pharmalyte pH 3-10"] will be set as the ampholyte in this standard.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or {Automatic, Null} entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Sample], Object[Sample]}] | _String) | {((ObjectP[{Model[Sample], Object[Sample]}] | _String) | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: Standards
StandardAmpholyteTargetConcentrations
The concentration (Vol/Vol) of amphoteric molecules in the master mix. When specifying values for each of Standards, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When StandardPremadeMasterMix is set to False, StandardAmpholyteTargetConcentrations is set to Automatic, and IncludeStandards is True, StandardAmpholyteTargetConcentrations will be set to 4% if one ampholyte is given, and 2% each, if more than one ampholyte is set and no StandardAmpholyteVolume values are given. If volume is given concentration will be calculated considering Ampholyte concentrations as 100%.
Pattern Description: Greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or list of one or more {Automatic, Null} or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent entries or Null.
Programmatic Pattern: ((RangeP[0.001*VolumePercent, 100*VolumePercent] | {(RangeP[0.001*VolumePercent, 100*VolumePercent] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: Standards
StandardAmpholyteVolume
The volume of amphoteric molecule stocks to add to the master mix. When specifying values for each of Standards, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When StandardAmpholyteVolume is set to Automatic, StandardPremadeMasterMix is set to False, and IncludeStandards is True, StandardAmpholyteVolume is set to the volume needed to reach the given AmpholyteTargetConcentrations based in the TotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or list of one or more {Automatic, Null} or greater than or equal to 0.1 microliters and less than or equal to 200 microliters entries or Null.
Programmatic Pattern: ((RangeP[0.1*Microliter, 200*Microliter] | {(RangeP[0.1*Microliter, 200*Microliter] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: Standards
StandardIsoelectricPointMarkers
Reference analytes, usually peptides with known isoelectric points, that are included in the master mix and are used to convert position in the capillary to the pI it represents. The mastermix for each sample should have two isoelectric point markers. A linear fit is then used to directly interpret the pI for each position between the two markers. When specifying values for each of Standards, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When StandardIsoelectricPointMarkers is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, {Model[Sample,StockSolution, "Resuspended cIEF pI Marker - 4.05"], Model[Sample,StockSolution, "Resuspended cIEF pI Marker - 9.99"]} will be set as the StandardIsoelectricPointMarkers in this standard.
Pattern Description: List of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample entries or list of one or more {Automatic, Null} entries or Null.
Programmatic Pattern: (({(ObjectP[{Model[Sample], Object[Sample]}] | _String)..} | {(Automatic | Null)..}) | Automatic) | Null
Index Matches to: Standards
StandardIsoelectricPointMarkersTargetConcentrations
The final concentration (Vol/Vol) of pI markers in the assay tube. When specifying values for each of Standards, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When StandardIsoelectricPointMarkersTargetConcentrations is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, pI markers target concentration will be set to 1%.
Pattern Description: Greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or list of one or more {Automatic, Null} or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent entries or Null.
Programmatic Pattern: ((RangeP[0.001*VolumePercent, 100*VolumePercent] | {(RangeP[0.001*VolumePercent, 100*VolumePercent] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: Standards
StandardIsoelectricPointMarkersVolume
The volume of pI marker stocks to add to the master mix. When specifying values for each of Standards, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When StandardIsoelectricPointMarkersVolume is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, it is set to the volume required to reach StandardIsoelectricPointMarkersTargetConcentrations in StandardTotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or list of one or more {Automatic, Null} or greater than or equal to 0.1 microliters and less than or equal to 200 microliters entries or Null.
Programmatic Pattern: ((RangeP[0.1*Microliter, 200*Microliter] | {(RangeP[0.1*Microliter, 200*Microliter] | (Automatic | Null))..}) | Automatic) | Null
Index Matches to: Standards
StandardElectroosmoticFlowBlocker
Electroosmotic flow blocker solution, usually methyl cellulose, to include in the master mix. Electroosmotic flow referes to the motion of liquids in a capillary as a result of applied voltage across it. Electroosmotic flow is most significant when channel diameters are very small. In capillary isoelectric focusing, the charge of the capillary wall should be masked to minimize the electroosmotic flow.
Default Calculation: When StandardElectroosmoticFlowBlocker is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, StandardElectroosmoticFlowBlocker will be set to Model[Sample,"1% Methyl Cellulose"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardElectroosmoticFlowBlockerTargetConcentrations
The concentration of ElectroosmoticFlowBlocker in the master mix.
Default Calculation: When StandardElectroosmoticFlowBlockerTargetConcentrations is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, it is set to 0.35% if no StandardElectroosmoticFlowBlockerVolume is set.
Pattern Description: Greater than or equal to 0.001 MassPercent and less than or equal to 100 MassPercent or Null.
Programmatic Pattern: (RangeP[0.001*MassPercent, 100*MassPercent] | Automatic) | Null
Index Matches to: Standards
StandardElectroosmoticFlowBlockerVolume
The volume of ElectroosmoticBlocker stock to add to the master mix.
Default Calculation: When StandardElectroosmoticFlowBlockerVolume is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, it is calculated based on StandardTotalVolume and the given StandardElectroosmoticFlowBlockerTargetConcentrations.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardDenature
Indicates if a DenaturationReagent should be added to the master mix.
Default Calculation: When StandardDenature is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, StandardDenature will be set to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standards
StandardDenaturationReagent
The denaturing agent, e.g., Urea or SimpleSol, to be added to the master mix to prevent protein precipitation.
Default Calculation: When StandardDenaturationReagent is set to Automatic, StandardPremadeMasterMix is False, StandardDenature is True, and IncludeStandards is True, StandardDenaturationReagent will be set to Model[Sample,StockSolution,"8M Urea"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardDenaturationReagentTargetConcentration
The final concentration of the denaturing agent in the master mix.
Default Calculation: When StandardDenaturationReagentTargetConcentration is set to Automatic, StandardPremadeMasterMix is False, StandardDenature is True, and IncludeStandards is True, it is set to 4M if no StandardDenaturationReagentVolume value is given. If StandardDenaturationReagentVolume is set and the concentration of StandardDenaturationReagent is known, StandardDenaturationReagentTargetConcentration is calculated.
Pattern Description: Greater than 0 molar or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | RangeP[0.001*VolumePercent, 100*VolumePercent]) | Automatic) | Null
Index Matches to: Standards
StandardDenaturationReagentVolume
The volume of the denaturing agent required to reach its final concentration in the master mix.
Default Calculation: When StandardDenaturationReagentVolume is set to Automatic, StandardPremadeMasterMix is False, StandardDenature is True, and IncludeStandards is True, it is set to the volume required to reach StandardDenaturationReagentTargetConcentration.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardIncludeAnodicSpacer
Indicates if an anodic spacer should be added to the master mix. Both acidic and alkaline carrier ampholytes may be lost in the electrolyte reservoirs due to diffusion and isotachophoresis, decreasing the resolution and detection of proteins at the extremes of the pH gradient. To reduce the loss of ampholytes, Spacers (ampholytes with very low or very high pIs) can be added to buffer the loss of analytes of interest. Traditionally, Iminodiacetic acid (pI 2.2) is added as an AnodicSpacer.
Default Calculation: When StandardIncludeAnodicSpacer is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, StandardIncludeAnodicSpacer will be set to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standards
StandardAnodicSpacer
Acidic ampholyte to include in the master mix. When analyzing a protein with very acidic pI (e.g. Pepsin), one might choose to add an AnodicSpacer such as Arginine (pI 10.7), to prevent loss of signal to the anolyte reservoir.
Default Calculation: When StandardAnodicSpacer is set to Automatic, StandardPremadeMasterMix is False, StandardIncludeAnodicSpacer is set to True, and IncludeStandards is True, StandardAnodicSpacer will be set to Model[Sample,StockSolution, "200mM Iminodiacetic acid"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardAnodicSpacerTargetConcentration
The final concentration of AnodicSpacer in the master mix.
Default Calculation: When StandardAnodicSpacerTargetConcentration is set to Automatic, StandardPremadeMasterMix is False, StandardIncludeAnodicSpacer is set to True, and IncludeStandards is True, it is set to 1mM when no StandardAnodicSpacerMarkersVolume is set. When StandardAnodicSpacer concentration is known and AnodicSpacerMarkersVolume is set, Automatic calculates the final concentration based on StandardAnodicSpacerTargetConcentrations and StandardTotalVolume.
Pattern Description: Greater than 0 millimolar or Null.
Programmatic Pattern: (GreaterP[0*Millimolar] | Automatic) | Null
Index Matches to: Standards
StandardAnodicSpacerVolume
The volume of AnodicSpacer stock to add to the master mix.
Default Calculation: When StandardAnodicSpacerVolume is set to Automatic, StandardPremadeMasterMix is False, StandardIncludeAnodicSpacer is set to True, and IncludeStandards is True, it is set to the volume required to reach StandardAnodicSpacerTargetConcentrations in StandardTotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standards
StandardIncludeCathodicSpacer
Indicates if a cathodic spacer should be added to the master mix. Both acidic and alkaline carrier ampholytes may be lost in the electrolyte reservoirs due to diffusion and isotachophoresis, decreasing the resolution and detection of proteins at the extremes of the pH gradient. To reduce the loss of ampholytes, Spacers (ampholytes with very low or very high pIs) can be added to buffer the loss of analytes of interest. Traditionally, Iminodiacetic acid (pI 2.2) and Arginine (pI 10.7) are added as Spacers.
Default Calculation: When StandardIncludeCathodicSpacer is set to Automatic, StandardPremadeMasterMix is False, and IncludeStandards is True, StandardIncludeCathodicSpacer will be set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standards
StandardCathodicSpacer
Basic ampholyte spacer to include in the master mix. When analyzing a protein with very basic pI (e.g. Cytochrome C), one might choose to add a CathodicSpacer such as Arginine (pI 10.7), to prevent loss of signal to the catholyte reservoir.
Default Calculation: When StandardCathodicSpacer is set to Automatic, StandardPremadeMasterMix is False, StandardIncludeCathodicSpacer is set to True, and IncludeStandards is True, StandardCathodicSpacer will be set to Model[Sample,StockSolution, "500mM Arginine"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standards
StandardCathodicSpacerTargetConcentration
The concentration of Cathodic spacer in the master mix.
Default Calculation: When StandardCathodicSpacerTargetConcentration is set to Automatic, StandardPremadeMasterMix is False, StandardIncludeCathodicSpacer is set to True, and IncludeStandards is True, it is set to 10 mM when no StandardCathodicSpacerMarkersVolume is set. When StandardCathodicSpacer concentration is known and StandardCathodicSpacerMarkersVolume is set, Automatic calculates the final concentration based on StandardCathodicSpacerTargetConcentrations and StandardTotalVolume.
Pattern Description: Greater than 0 millimolar or Null.
Programmatic Pattern: (GreaterP[0*Millimolar] | Automatic) | Null
Index Matches to: Standards
StandardCathodicSpacerVolume
The volume of Cathodic spacer stocks to add to the master mix.
Default Calculation: When StandardCathodicSpacerVolume is set to Automatic, StandardPremadeMasterMix is False, StandardIncludeCathodicSpacer is set to True, and IncludeStandards is True, it is set to the volume required to reach StandardAnodicSpacerTargetConcentrations in StandardTotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standards
Standard Separation
StandardLoadTime
Time to load standards in master mix into the capillary by vacuum.
Default Calculation: When StandardLoadTime is set to Automatic and IncludeStandards is True, StandardLoadTime is set to 55 seconds.
Pattern Description: Greater than or equal to 1 second and less than or equal to 300 seconds or Null.
Programmatic Pattern: (RangeP[1*Second, 300*Second] | Automatic) | Null
Index Matches to: Standards
StandardVoltageDurationProfile
Series of voltages and durations to apply onto the capillary for separation. Supports up to 20 steps where each step is 0-600 minutes, and 0-5000 volts.
Default Calculation: When StandardVoltageDurationProfile is set to Automatic and IncludeStandards is True, StandardVoltageDurationProfile is set to a two-step focusing: 1500 Volts for 1 Minute, followed by 3000 Volts for 10 minutes.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: ({{RangeP[0.1*Volt, 5000*Volt], RangeP[0.01*Minute, 600*Minute]}..} | Automatic) | Null
Index Matches to: Standards
Standard Detection
StandardImagingMethods
Whole capillary imaging by Either UVAbsorbance (280 nm) alone or both UVAbsorbance and Native Fluorescence (Ex 280 nm, Em 320-450nm).
Default Calculation: When StandardImagingMethods is set to Automatic and IncludeStandards is True, StandardImagingMethods is set to imaging by both UVAbsorbance and NativeFluorescence.
Pattern Description: Absorbance or AbsorbanceAndFluorescence or Null.
Programmatic Pattern: ((Absorbance | AbsorbanceAndFluorescence) | Automatic) | Null
Index Matches to: Standards
StandardNativeFluorescenceExposureTime
Exposure duration for NativeFluorescence detection.
Default Calculation: When StandardNativeFluorescenceExposureTime is set to Automatic and IncludeStandards is True, StandardNativeFluorescenceExposureTime is set to imaging for 3, 5, 10, and 20 seconds.
Pattern Description: Greater than or equal to 1 second and less than or equal to 200 seconds or list of one or more greater than or equal to 1 second and less than or equal to 200 seconds entries or Null.
Programmatic Pattern: ((RangeP[1*Second, 200*Second] | {RangeP[1*Second, 200*Second]..}) | Automatic) | Null
Index Matches to: Standards
Blanks
IncludeBlanks
Indicates if blanks should be included in this experiment. Blanks serve to identify any artifacts of sample prep that may affect the results of this experiment.
Default Calculation: When IncludeBlanks is set to Automatic and any of its related options is specified, it is set to True. Otherwise, it is set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Blanks
Indicates what blanks to include.
Default Calculation: When Blanks is set to Automatic and IncludeBlanks is True, it is set to Milli-Q water.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Blanks and Blanks
BlankVolume
Indicates the volume drawn from the blank to the assay tube. Each tube contains a Blank, ampholytes, pI markers, a reducing Agent, an electroosmotic flow blocker, and an anodic and/or cathodic spacers.
Default Calculation: When BlankVolume is set to Automatic and IncludeBlanks is True, the volume is calculated to be 10% of the BlankTotalVolume. When IncludeBlanks is False, BlankVolume is Null.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Blank Preparation
BlankFrequency
Indicates how many injections per Blank should be included in this experiment. Sample, Standard, and Blank injection order are resolved according to InjectionTable.
Default Calculation: When BlankFrequency is set to Automatic and IncludeBlanks is True, it will default to FirstAndLast.
Pattern Description: _?(GreaterQ[#1, 0, 1] & ), FirstAndLast, First, or Last or Null.
Programmatic Pattern: ((GreaterP[0, 1] | FirstAndLast | First | Last) | Automatic) | Null
BlankTotalVolume
Indicates the final volume in the assay tube, including blank and master mix.
Default Calculation: When BlankTotalVolume is set to Automatic and IncludeBlanks is True, it will resolve to the most common total volume in SamplesIn.
Pattern Description: Greater than or equal to 50 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[50*Microliter, 200*Microliter] | Automatic) | Null
BlankPremadeMasterMix
Indicates if a premade should be used or, alternatively, the master should be made as part of this protocol. The contains the reagents required for cIEF experiments, i.e., DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When BlankPremadeMasterMix is set to Automatic and IncludeBlanks is True, it will resolve to True if any of its downstream options is specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankPremadeMasterMixReagent
The premade master mix used for cIEF experiment, containing DenaturationReagent, Ampholytes, IsoelectricPointMarkers, Spacers, and ElectroosmoticBlocker.
Default Calculation: When BlankPremadeMasterMix is True and BlankPremadeMasterMixReagent is set to Automatic, it will resolve to Model[Sample, StockSolution, "2X Wide-Range cIEF Premade Master Mix"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankPremadeMasterMixDiluent
The solution used to dilute the premade master mix used to its working concentration.
Default Calculation: When BlankPremadeMasterMixDiluent is set to Automatic and BlankPremadeMasterMix is set to True, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankPremadeMasterMixReagentDilutionFactor
The factor by which the premade master mix should be diluted by in the final assay tube.
Default Calculation: When BlankPremadeMasterMixReagentDilutionFactor is set to Automatic and BlankPremadeMasterMix is set to True, it will be set as the ratio of the total volume to premade master mix volume.
Pattern Description: Greater than or equal to 1 or Null.
Programmatic Pattern: (GreaterEqualP[1] | Automatic) | Null
BlankPremadeMasterMixVolume
The volume of the premade master mix required to reach its final concentration.
Default Calculation: When BlankPremadeMasterMixVolume is set to Automatic and BlankPremadeMasterMix is set to True, the volume is calculated by the division of TotalVolume by PremadeMasterMixReagentDilutionFactor. If PremadeMasterMix is set to False, PremadeMasterMixVolume is Null.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
BlankDiluent
The solution used to top volume in assay tube to total volume and dilute components to working concentration.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankDiluent is set to Automatic, Model[Sample,"Milli-Q water"] will be set as diluent.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankAmpholytes
A solution composed of a mixture of amphoteric molecules, that case act both as an acid and a base, in the master mix that form the pH gradient. In the presence of an anolyte and a catholyte, and once a voltage is applied across the capillary, ampholytes for a pH gradient at a range that depend on the amopholyte composition. Proteins then resolve according to their isoelectric point within this gradient. When specifying values for each of Blanks, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When BlankAmpholytes is set to Automatic and BlankPremadeMasterMix is False, Model[Sample,"Pharmalyte pH 3-10"] will be set as the ampholyte in this standard.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or list of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or {Automatic, Null} entries or Null.
Programmatic Pattern: (((ObjectP[{Model[Sample], Object[Sample]}] | _String) | {((ObjectP[{Model[Sample], Object[Sample]}] | _String) | (Automatic | Null))..}) | Automatic) | Null
BlankAmpholyteTargetConcentrations
The concentration (Vol/Vol) of amphoteric molecules in the master mix. When specifying values for each of Blanks, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When BlankPremadeMasterMix is set to False and BlankAmpholyteTargetConcentrations is set to Automatic, AmpholyteTargetConcentrations will be set to 4% if one ampholyte is given, and 2% each, if more than one ampholyte is set and no AmpholyteVolume values are given. If volume is given concentration will be calculated considering Ampholyte concentrations as 100%.
Pattern Description: Greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or list of one or more {Automatic, Null} or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent entries or Null.
Programmatic Pattern: ((RangeP[0.001*VolumePercent, 100*VolumePercent] | {(RangeP[0.001*VolumePercent, 100*VolumePercent] | (Automatic | Null))..}) | Automatic) | Null
BlankAmpholyteVolume
The volume of amphoteric molecule stocks to add to the master mix. When specifying values for each of Blanks, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When BlankAmpholyteVolume is set to Automatic and BlankPremadeMasterMix is set to False, BlankAmpholyteVolume is set to the volume needed to reach the given AmpholyteTargetConcentrations based in the TotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or list of one or more {Automatic, Null} or greater than or equal to 0.1 microliters and less than or equal to 200 microliters entries or Null.
Programmatic Pattern: ((RangeP[0.1*Microliter, 200*Microliter] | {(RangeP[0.1*Microliter, 200*Microliter] | (Automatic | Null))..}) | Automatic) | Null
BlankIsoelectricPointMarkers
Reference analytes, usually peptides with known isoelectric points, that are included in the master mix and are used to convert position in the capillary to the pI it represents. The mastermix for each sample should have two isoelectric point markers. A linear fit is then used to directly interpret the pI for each position between the two markers. When specifying values for each of Blanks, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When BlankIsoelectricPointMarkers is set to Automatic and BlankPremadeMasterMix is False, {Model[Sample,StockSolution, "Resuspended cIEF pI Marker - 4.05"], Model[Sample,StockSolution, "Resuspended cIEF pI Marker - 9.99"]} will be set as the BlankIsoelectricPointMarkers in this standard.
Pattern Description: List of one or more an object of type or subtype Model[Sample] or Object[Sample] or a prepared sample entries or list of one or more {Automatic, Null} entries or Null.
Programmatic Pattern: (({(ObjectP[{Model[Sample], Object[Sample]}] | _String)..} | {(Automatic | Null)..}) | Automatic) | Null
BlankIsoelectricPointMarkersTargetConcentrations
The final concentration (Vol/Vol) of pI markers in the assay tube. When specifying values for each of Blanks, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When BlankIsoelectricPointMarkersTargetConcentrations is set to Automatic and BlankPremadeMasterMix is False, pI markers target concentration will be set to 1%.
Pattern Description: Greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or list of one or more {Automatic, Null} or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent entries or Null.
Programmatic Pattern: ((RangeP[0.001*VolumePercent, 100*VolumePercent] | {(RangeP[0.001*VolumePercent, 100*VolumePercent] | (Automatic | Null))..}) | Automatic) | Null
BlankIsoelectricPointMarkersVolume
The volume of pI marker stocks to add to the master mix. When specifying values for each of Blanks, please specify this option as a list of lists to avoid ambiguity.
Default Calculation: When BlankIsoelectricPointMarkersVolume is set to Automatic and BlankPremadeMasterMix is False, it is set to the volume required to reach BlankIsoelectricPointMarkersTargetConcentrations in BlankTotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or list of one or more {Automatic, Null} or greater than or equal to 0.1 microliters and less than or equal to 200 microliters entries or Null.
Programmatic Pattern: ((RangeP[0.1*Microliter, 200*Microliter] | {(RangeP[0.1*Microliter, 200*Microliter] | (Automatic | Null))..}) | Automatic) | Null
BlankElectroosmoticFlowBlocker
Electroosmotic flow blocker solution, usually methyl cellulose, to include in the master mix. Electroosmotic flow referes to the motion of liquids in a capillary as a result of applied voltage across it. Electroosmotic flow is most significant when channel diameters are very small. In capillary isoelectric focusing, the charge of the capillary wall should be masked to minimize the electroosmotic flow.
Default Calculation: When BlankElectroosmoticFlowBlocker is set to Automatic and BlankPremadeMasterMix is False, BlankElectroosmoticFlowBlocker will be set to Model[Sample,"1% Methyl Cellulose"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankElectroosmoticFlowBlockerTargetConcentrations
The concentration of ElectroosmoticFlowBlocker in the master mix.
Default Calculation: When BlankElectroosmoticFlowBlockerTargetConcentrations is set to Automatic and BlankPremadeMasterMix is False, it is set to 0.35% if no BlankElectroosmoticFlowBlockerVolume is set.
Pattern Description: Greater than or equal to 0.001 MassPercent and less than or equal to 100 MassPercent or Null.
Programmatic Pattern: (RangeP[0.001*MassPercent, 100*MassPercent] | Automatic) | Null
BlankElectroosmoticFlowBlockerVolume
The volume of ElectroosmoticBlocker stock to add to the master mix.
Default Calculation: When BlankElectroosmoticFlowBlockerVolume is set to Automatic and BlankPremadeMasterMix is False, it is calculated based on BlankTotalVolume and the given BlankElectroosmoticFlowBlockerTargetConcentrations.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
BlankDenature
Indicates if a DenaturationReagent should be added to the master mix.
Default Calculation: When BlankDenature is set to Automatic and BlankPremadeMasterMix is False, BlankDenature will be set to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankDenaturationReagent
The denaturing agent, e.g., Urea or SimpleSol, to be added to the master mix to prevent protein precipitation.
Default Calculation: When BlankDenaturationReagent is set to Automatic, BlankPremadeMasterMix is False, and BlankDenature is True, BlankDenaturationReagent will be set to Model[Sample,StockSolution,"8M Urea"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankDenaturationReagentTargetConcentration
The final concentration of the denaturing agent in the master mix.
Default Calculation: When BlankDenaturationReagentTargetConcentration is set to Automatic, BlankPremadeMasterMix is False, and BlankDenature is True, it is set to 4M if no BlankDenaturationReagentVolume value is given. If BlankDenaturationReagentVolume is set and the concentration of BlankDenaturationReagent is known, BlankDenaturationReagentTargetConcentration is calculated.
Pattern Description: Greater than 0 molar or greater than or equal to 0.001 VolumePercent and less than or equal to 100 VolumePercent or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | RangeP[0.001*VolumePercent, 100*VolumePercent]) | Automatic) | Null
BlankDenaturationReagentVolume
The volume of the denaturing agent required to reach its final concentration in the master mix.
Default Calculation: When BlankDenaturationReagentVolume is set to Automatic, BlankPremadeMasterMix is False, and BlankDenature is True, it is set to the volume required to reach BlankDenaturationReagentTargetConcentration.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
BlankIncludeAnodicSpacer
Indicates if an anodic spacer should be added to the master mix. Both acidic and alkaline carrier ampholytes may be lost in the electrolyte reservoirs due to diffusion and isotachophoresis, decreasing the resolution and detection of proteins at the extremes of the pH gradient. To reduce the loss of ampholytes, Spacers (ampholytes with very low or very high pIs) can be added to buffer the loss of analytes of interest. Traditionally, Iminodiacetic acid (pI 2.2) is added as an AnodicSpacer.
Default Calculation: When BlankIncludeAnodicSpacer is set to Automatic and BlankPremadeMasterMix is False, BlankIncludeAnodicSpacer will be set to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankAnodicSpacer
Acidic ampholyte to include in the master mix. When analyzing a protein with very acidic pI (e.g. Pepsin), one might choose to add an AnodicSpacer such as Arginine (pI 10.7), to prevent loss of signal to the anolyte reservoir.
Default Calculation: When BlankAnodicSpacer is set to Automatic, BlankPremadeMasterMix is False and BlankIncludeAnodicSpacer is set to True, BlankAnodicSpacer will be set to Model[Sample,StockSolution, "200mM Iminodiacetic acid"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankAnodicSpacerTargetConcentration
The final concentration of AnodicSpacer in the master mix.
Default Calculation: When BlankAnodicSpacerTargetConcentration is set to Automatic, BlankPremadeMasterMix is False and BlankIncludeAnodicSpacer is set to True, it is set to 1mM when no BlankAnodicSpacerMarkersVolume is set. When BlankAnodicSpacer concentration is known and AnodicSpacerMarkersVolume is set, Automatic calculates the final concentration based on BlankAnodicSpacerTargetConcentrations and BlankTotalVolume.
Pattern Description: Greater than 0 millimolar or Null.
Programmatic Pattern: (GreaterP[0*Millimolar] | Automatic) | Null
BlankAnodicSpacerVolume
The volume of AnodicSpacer stock to add to the master mix.
Default Calculation: When BlankAnodicSpacerVolume is set to Automatic, BlankPremadeMasterMix is False and BlankIncludeAnodicSpacer is set to True, it is set to the volume required to reach BlankAnodicSpacerTargetConcentrations in BlankTotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
BlankIncludeCathodicSpacer
Indicates if a cathodic spacer should be added to the master mix. Both acidic and alkaline carrier ampholytes may be lost in the electrolyte reservoirs due to diffusion and isotachophoresis, decreasing the resolution and detection of proteins at the extremes of the pH gradient. To reduce the loss of ampholytes, Spacers (ampholytes with very low or very high pIs) can be added to buffer the loss of analytes of interest. Traditionally, Iminodiacetic acid (pI 2.2) and Arginine (pI 10.7) are added as Spacers.
Default Calculation: When BlankIncludeCathodicSpacer is set to Automatic and BlankPremadeMasterMix is False, BlankIncludeCathodicSpacer will be set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankCathodicSpacer
Basic ampholyte spacer to include in the master mix. When analyzing a protein with very basic pI (e.g. Cytochrome C), one might choose to add a CathodicSpacer such as Arginine (pI 10.7), to prevent loss of signal to the catholyte reservoir.
Default Calculation: When BlankCathodicSpacer is set to Automatic, BlankPremadeMasterMix is False and BlankIncludeCathodicSpacer is set to True, BlankCathodicSpacer will be set to Model[Sample,StockSolution, "500mM Arginine"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankCathodicSpacerTargetConcentration
The concentration of Cathodic spacer in the master mix.
Default Calculation: When BlankCathodicSpacerTargetConcentration is set to Automatic, BlankPremadeMasterMix is False and BlankIncludeCathodicSpacer is set to True, it is set to 10 mM when no BlankCathodicSpacerMarkersVolume is set. When BlankCathodicSpacer concentration is known and BlankCathodicSpacerMarkersVolume is set, Automatic calculates the final concentration based on BlankCathodicSpacerTargetConcentrations and BlankTotalVolume.
Pattern Description: Greater than 0 millimolar or Null.
Programmatic Pattern: (GreaterP[0*Millimolar] | Automatic) | Null
BlankCathodicSpacerVolume
The volume of Cathodic spacer stocks to add to the master mix.
Default Calculation: When BlankCathodicSpacerVolume is set to Automatic, BlankPremadeMasterMix is False and BlankIncludeCathodicSpacer is set to True, it is set to the volume required to reach BlankAnodicSpacerTargetConcentrations in BlankTotalVolume.
Pattern Description: Greater than or equal to 0.1 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0.1*Microliter, 200*Microliter] | Automatic) | Null
Blank Separation
BlankLoadTime
Time to load standards in master mix into the capillary by vacuum.
Default Calculation: When BlankLoadTime is set to Automatic and IncludeBlanks is True, BlankLoadTime is set to 55 seconds.
Pattern Description: Greater than or equal to 1 second and less than or equal to 300 seconds or Null.
Programmatic Pattern: (RangeP[1*Second, 300*Second] | Automatic) | Null
BlankVoltageDurationProfile
Series of voltages and durations to apply onto the capillary for separation. Supports up to 20 steps where each step is 0-600 minutes, and 0-5000 volts.
Default Calculation: When BlankVoltageDurationProfile is set to Automatic and IncludeBlanks is True, BlankVoltageDurationProfile is set to a two-step focusing: 1500 Volts for 1 Minute, followed by 3000 Volts for 10 minutes.
Pattern Description: List of one or more {Voltage, Time} entries or Null.
Programmatic Pattern: ({{RangeP[0.001*Volt, 5000*Volt], RangeP[0.01*Minute, 600*Minute]}..} | Automatic) | Null
BlankImagingMethods
Whole capillary imaging by Either UVAbsorbance (280 nm) alone or both UVAbsorbance and Native Fluorescence (Ex 280 nm, Em 320-450nm).
Default Calculation: When BlankImagingMethods is set to Automatic and IncludeBlanks is True, BlankImagingMethods is set to imaging by both UVAbsorbance and NativeFluorescence.
Pattern Description: Absorbance or AbsorbanceAndFluorescence or Null.
Programmatic Pattern: ((Absorbance | AbsorbanceAndFluorescence) | Automatic) | Null
BlankNativeFluorescenceExposureTime
Exposure duration for NativeFluorescence detection.
Default Calculation: When BlankNativeFluorescenceExposureTime is set to Automatic and IncludeBlanks is True, BlankNativeFluorescenceExposureTime is set to imaging for 3, 5, 10, and 20 seconds.
Pattern Description: Greater than or equal to 1 second and less than or equal to 200 seconds or list of one or more greater than or equal to 1 second and less than or equal to 200 seconds entries or Null.
Programmatic Pattern: ((RangeP[1*Second, 200*Second] | {RangeP[1*Second, 200*Second]..}) | Automatic) | Null
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and when the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise set to Model[Container, Vessel, "2mL Tube"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[Model[Container]] | Automatic) | Null
Index Matches to: experiment samples
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
Index Matches to: experiment samples