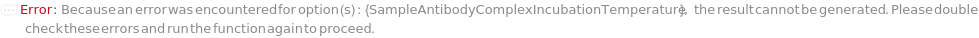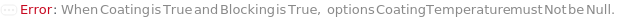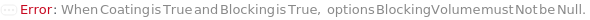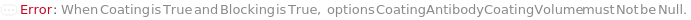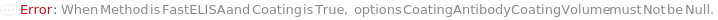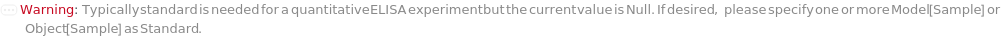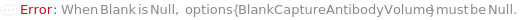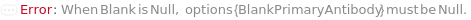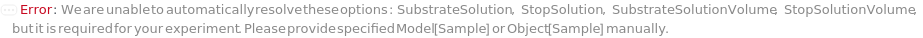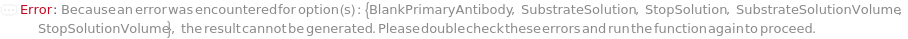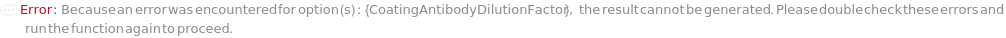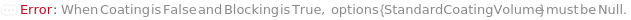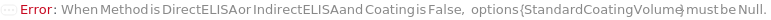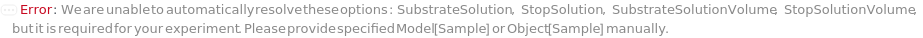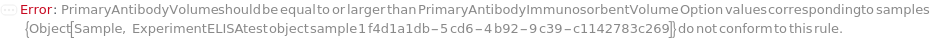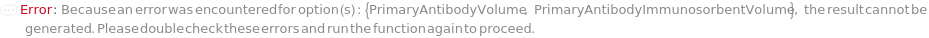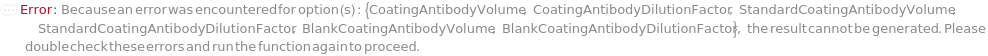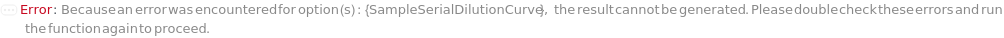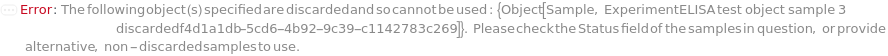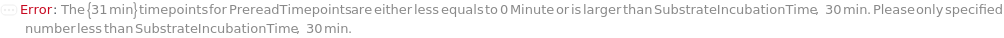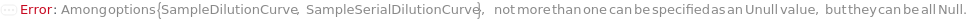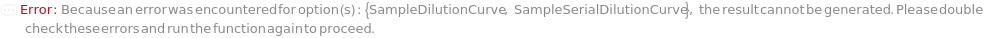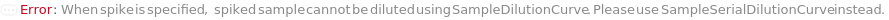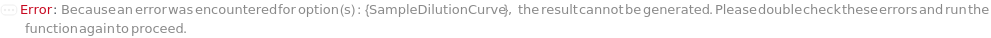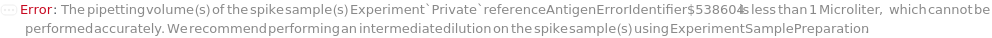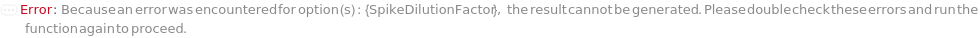General
Method
Defines the type of ELISA experiment to be performed. Types include DirectELISA, IndirectELISA, DirectSandwichELISA, IndirectSandwichELISA, DirectCompetitiveELISA, IndirectCompetitiveELISA, and FastELISA. Compared with the Direct methods where the primary antibody is conjugated with an enzyme (such as Horse Radish Peroxidase/HRP or Alkaline Phosphatas/AP) for detection, in all Indirect methods a secondary antibody is, instead, conjugated with this enzyme. In Indirect methods, an additional step of SecondaryAntibody incubation is added to the corresponding Direct methods. In a DirectELISA experiment, the Sample is coated on the ELISAPlate, and then a primary antibody is used to detect the target antigen. In a DirectSandwichELISA experiment, a capture antibody is coated onto the ELISAPlate to pull down the target antigen from the sample. Then a primary antibody is used to detect the target antigen. In a DirectCompetitiveELISA experiment, a reference antigen is used to coat the ELISAPlate. Samples are incubated with the primary antibody. Then, when the Sample-Antibody-Complex solution is loaded on the ELISAPlate, the remaining free primary antibody binds to the reference antigen. In a FastELISA experiment, a coating antibody against a tag is coated to the ELISAPlate. A capture antibody containing this tag and a primary antibody for antigen detection is incubated with the sample to form a CaptureAntibody-TargetAntigen-PrimaryAntibody complex. Then this complex is pulled down by the coating antibody to the surface of the plate (See Figure 1.1).
Default Value: DirectELISA
Pattern Description: DirectELISA, IndirectELISA, DirectSandwichELISA, IndirectSandwichELISA, DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA.
Programmatic Pattern: ELISAMethodP
TargetAntigen
The Analyte molecule (e.g., peptide, protein, and hormone) detected and quantified in the samples by Antibodies in the ELISA experiment. This option is used to automatically set sample Antibodies and the corresponding experiment conditions of Standards and Blanks.
Default Calculation: Automatically set to the Model[Molecule] in the Analyte field of the sample.
Pattern Description: An object of type or subtype Model[Molecule] or Null.
Programmatic Pattern: (ObjectP[{Model[Molecule]}] | Automatic) | Null
Index Matches to: experiment samples
NumberOfReplicates
The number of times an ELISA assay will be repeated in parallel wells. Samples, Standards, or Blanks will be loaded this number of times in the plates. Replications are conducted at the same time, and if possible, in the same ELISAPlate. If set to Null, each Sample, Standard, or Blank will be assayed once.
Default Calculation: Automatically set to 2 unless method is DirectELISA or IndirectELISA and Coating is False, in which case it is set to Null.
Pattern Description: Greater than or equal to 2 and less than or equal to 192 in increments of 1 or Null.
Programmatic Pattern: (RangeP[2, 192, 1] | Automatic) | Null
WashingBuffer
The solution used to rinse off unbound molecules from the assay plate.
Default Value: Model[Sample, StockSolution, Phosphate Buffered Saline with 0.05% TWEEN 20, pH 7.4]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
Sample Assembly
Spike
The sample with a known concentration of analyte. Spike is to be mixed with the input sample. The purpose of spiking is to perform a spike-and-recovery assessment to determine whether the ELISA can accurately test the concentration of analyte within the context of the sample. If the recovery observed for the spiked sample is identical to the analyte prepared in standard diluent, the sample is considered not to interfere with the assay. For example, if molecules in the sample inhibits the binding between the antibody and TargetAntigen, the results from the ELISA becomes inaccurate. In a Spike-and-Recovery experiment,an aliquot of a sample is mixed with Spike. ELISA is performed on the Sample alone (also called Neat Sample) and the Spiked Sample at the same time, where the Spike concentration in the sample can be measured. This measured Spike concentration can be then compared with the known Spike concentration. Typically a 20% difference between measured Spike concentration and the known Spike concentration is acceptable. Spiked sample can be further diluted to perform linearity-of-dilution assessment. The plate used for sample spiking and dilution will be discarded after the experiment.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null
Index Matches to: experiment samples
SpikeDilutionFactor
The dilution ratio by which the Spike is mixed with the input sample before further dilution is performed.
Default Calculation: Automatically set to 0.1 if Spike is not Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: experiment samples
SpikeStorageCondition
The condition under which the unused portion of spike should be stored.
Default Calculation: Automatically set to Refrigerator if Spike is not Null.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
SampleSerialDilutionCurve
The collection of serial dilutions that will be performed on each sample: if spiking was performed, the dilution will be performed on the spiked sample; otherwise, the dilutions will be performed on the sample. This Linearity-of-Dilution step assesses whether the ELISA experiment can reliably measure TargetAntigen concentration at different concentration ranges. For example, a sample with TargetAntigen concentration of 100ng/ul, if diluted 1:2, 1:4, and 1:8, should yield ELISA measurements of 50ng/ul, 25ng/ul, and 12.5ng/ul (or should follow a concentration ratio of 4:2:1). Typically a 20% difference between the measured and expected concentrations is considered acceptable. For Serial Dilution Volumes, the Transfer Volume is taken out of the sample and added to a second well with the Diluent Volume of the Diluent. It is mixed, then the Transfer Volume is taken out of that well to be added to a third well. This is repeated to make Number Of Dilutions diluted samples. For example, if a 100 ug/ ml sample with a Transfer Volume of 20 Microliters, a Diluent Volume of 60 Microliters and a Number of Dilutions of 3 is used, it will create a DilutionCurve of 25 ug/ ml, 6.25 ug/ ml, and 1.5625 ug/ ml with each dilution having a volume of 60 Microliters. For Serial Dilution Factors, the sample will be diluted by the dilution factor at each transfer step. IMPORTANT: because the dilution curve does not intrinsically include the original sample, in the case of sample dilution the first diluting factor should be 1 or Diluent Volume should be 0 Microliter to include the original sample. For example, if a Spike-and-Recovery assay is to be performed, the first dilution factor must be 1 in order to include the original spiked sample. During experiment, an 5% extra volume than specified is going to be prepared in order to offset pipetting errors. Therefore, the total volume for each well specified in this option should be equal or greater than the volume needed for the experiment. Because the dilution of each assay (well) is prepared independently, the Number of Replications should not be considered when determining the Spike Volume. The plate used for sample dilutions will be discarded after the experiment. Note: if spike or antibodies are to be mixed with samples, their volumes are counted towards diluent volume.
Default Calculation: The option is automatically set Null if or a SampleDilutionCurve is specified.
Pattern Description: Serial Dilution Factor or Serial Dilution Volumes or Null.
Programmatic Pattern: (({RangeP[10*Microliter, 1500*Milliliter], {RangeP[0, 1], GreaterP[0, 1]} | {RangeP[0, 1]..}} | {RangeP[0*Microliter, 1000*Microliter], RangeP[0*Microliter, 1000*Microliter], GreaterP[0, 1]}) | Automatic) | Null
Index Matches to: experiment samples
SampleDilutionCurve
The collection of dilutions that will be performed on each sample: the dilutions will be performed on the sample. Spiked samples should only be further diluted with serial dilution. This Linearity-of-Dilution step assesses whether the ELISA experiment can reliably measure TargetAntigen concentration at different concentration ranges. For example, a sample with TargetAntigen concentration of 100ng/ul, if diluted 1:2, 1:4, and 1:8, should yield ELISA measurements of 50ng/ul, 25ng/ul, and 12.5ng/ul (or should follow a concentration ratio of 8:4:2:1). Typically a 20% difference between the measured and expected concentrations is considered acceptable. Linearity-of-Dilution experiment can be done with Spiked or non-Spiked samples. For Fixed Dilution Volume Dilution Curves, the Spike Amount is the volume of the sample that will be mixed the Diluent Volume of the Diluent to create a desired concentration. The Assay Volume is the TOTAL volume of the sample that will be created after being diluted by the Dilution Factor. IMPORTANT: because the dilution curve does not intrinsically include the original sample, in the case of sample dilution the first diluting factor must be 1, or Diluent Volume should be 0 to include the original sample. During experiment, an 5% extra volume than specified is going to be prepared in order to offset pipetting errors. Therefore, the total volume for each well specified in this option should be equal or greater than the volume needed for the experiment. Because the dilution of each assay (well) is prepared independently, the Number of Replications should not be considered when determining the Spike Volume. The plate used for sample dilutions will be discarded after the experiment. Note: if spike or antibodies are to be mixed with samples, their volumes are counted towards diluent volume.
Default Calculation: The option is automatically set Null if or a SampleSerialDilutionCurve is specified.
Pattern Description: Fixed Dilution Factor or Fixed Dilution Volume or Null.
Programmatic Pattern: (({{RangeP[10*Microliter, 1500*Microliter], RangeP[0, 1]}..} | {{RangeP[0*Microliter, 1000*Microliter], RangeP[0*Microliter, 1000*Microliter]}..}) | Automatic) | Null
Index Matches to: experiment samples
SampleDiluent
The buffer used to perform multiple dilutions of samples or spiked samples.
Default Calculation: If SampleDilutionCurve and SampleSerialDilutionCurve are not both Null, in the case when Method is set to DirectELISA and IndirectELISA, the option is automatically set to Model[Sample, StockSolution, "1x Carbonate-Bicarbonate Buffer pH10"]; in all other cases, the option is automatically set to Model[Sample,"ELISA Blocker Blocking Buffer"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Antibody Antigen Preparation
CoatingAntibody
The sample containing the antibody that is used for coating in FastELISA.
Default Calculation: If Method is FastELISA, automatically set to an antibody against a tag which is conjugated to a the CaptureAntibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CoatingAntibodyDilutionFactor
The dilution ratio of CoatingAntibody. CoatingAntibody is diluted with CoatingAntibodyDiluent. Either CoatingAntibodyDilutionFactor or CoatingAntibodyVolume should be provided but not both.
Default Calculation: Automatically set to 0.001 (1:1,000) for FastELISA.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: experiment samples
CoatingAntibodyVolume
The volume of undiluted CoatingAntibody added into the corresponding well of the assay plate. CoatingAntibody is diluted with CoatingAntibodyDiluent. CoatingAntibodyVolume is used as an alternative to CoatingAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: experiment samples
CoatingAntibodyDiluent
The buffer used to dilute CoatingAntibodies.
Default Calculation: If Method is set to FastELISA, the option is automatically set to Model[Sample, StockSolution, "1x Carbonate-Bicarbonate Buffer pH10"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
CoatingAntibodyStorageCondition
The condition under which the unused portion of Coating Antibody stock sample should be stored.
Default Calculation: Automatically set to Refrigerator if Method is set to FastELISA.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibody
The sample containing the antibody that is used to pull down the antigen from sample solution to the surface of the assay plate wells in DirectSandwichELISA, IndirectSandwichELISA, and FastELISA.
Default Calculation: If Method is FastELISA, automatically set to an antibody containing an affinity tag and against the TargetAntigen. If Method is DirectSandwichELISA or IndirectSandwichELISA, automatically set to an un-tagged antibody against TargetAntigen.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyDilutionFactor
The dilution ratio of CaptureAntibody. For DirectSandwichELISA and IndirectSandwichELISA, CaptureAntibody is diluted with CaptureAntibodyDiluent. For FastELISA, CaptureAntibody is diluted in the corresponding sample. Either CaptureAntibodyDilutionFactor or CaptureAntibodyVolume should be provided but not both.
Default Calculation: For DirectSandwichELISA and IndirectSandwichELISA, automatically set to 0.001 (1:1,000). For FastELISA, automatically set to 0.01 (1:100).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyVolume
The volume of undiluted CaptureAntibody added into the corresponding well of the assay plate. CaptureAntibodyVolume is used as an alternative to CaptureAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: experiment samples
CaptureAntibodyDiluent
The buffer used to dilute CaptureAntibodies. Set to Null when CapturaAntibody should be diluted into samples (in FastELISA).
Default Calculation: If Method is set to DirectSandwichELISA or IndirectSandwichELISA, the option is automatically set to Model[Sample, StockSolution, "1x Carbonate-Bicarbonate Buffer pH10"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
CaptureAntibodyStorageCondition
The condition under which the unused portion of Capture Antibody stock sample should be stored.
Default Calculation: If Method is set to DirectSandwichELISA, IndirectSandwichELISA, or FastELISA, Automatically set to Refrigerator.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
ReferenceAntigen
The sample containing the antigen that is used in DirectCompetitiveELISA or IndirectCompetitiveELISA. The ReferenceAntigen competes with TargetAntigen in the samples for the binding of the PrimaryAntibody. Reference Antigen is also referred to as Inhibitor Antigen.
Default Calculation: Automatically set to a sample containing known amount of TargetAntigen when Method is set to DirectCompetitiveELISA or IndirectCompetitiveELISA.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
ReferenceAntigenDilutionFactor
The dilution ratio of ReferenceAntigen. The ReferenceAntigenSample is always diluted in ReferenceAntigenDiluent. Either ReferenceAntigenDilutionFactor or ReferenceAntigenVolume should be provided but not both.
Default Calculation: If Method is DirectCompetitiveELISA or IndirectCompetitiveELISA, automatically set to 0.001 (1:1,000).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: experiment samples
ReferenceAntigenVolume
The volume of undiluted ReferenceAntigen added into the corresponding well of the assay plate. ReferenceAntigenVolume is used as an alternative to ReferenceAntigenDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: experiment samples
ReferenceAntigenDiluent
The buffer used to dilute the ReferenceAntigen.
Default Calculation: Method is DirectCompetitiveELISA and IndirectCompetitiveELISA, the option is automatically set to Model[Sample, StockSolution, "1x Carbonate-Bicarbonate Buffer pH10"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
ReferenceAntigenStorageCondition
The condition under which the unused portion of Reference Antigen stock sample should be stored.
Default Calculation: Automatically set to Refrigerator if Method is DirectCompetitiveELISA and IndirectCompetitiveELISA.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
PrimaryAntibody
The antibody that directly binds with the TargetAntigen (analyte).
Default Calculation: The option will be automatically set to an antibody against the TargetAntigen.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
PrimaryAntibodyDilutionFactor
The dilution ratio of PrimaryAntibody. For DirectELISA, IndirectELISA, DirectSandwichELISA, and IndirectSandwichELISA, the antibody is diluted with PrimaryAntibodyDiluent. For DirectCompetitiveELISA, IndirectCompetitiveELISA, and FastELISA, the antibody is diluted with the corresponding sample. Either PrimaryAntibodyDilutionFactor or PrimaryAntibodyVolume should be provided but not both.
Default Calculation: If used for DirectELISA, IndirectELISA, DirectSandwichELISA, IndirectSandwichELISA, automatically set to 0.001 (1:1,000). If used for DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, automatically set to 0.01 (1:100).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: experiment samples
PrimaryAntibodyVolume
The volume of undiluted PrimaryAntibody added into the corresponding well of the assay plate. PrimaryAntibodyVolume is used as an alternative to PrimaryAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: experiment samples
PrimaryAntibodyDiluent
The buffer used to dilute the PrimaryAntibody.Set to Null when PrimaryAntibody should be diluted into samples (in DirectCompetitiveELISA, IndirectCompetitiveELISA, and FastELISA).
Default Calculation: If Method is set to DirectELISA, IndirectELISA, DirectSandwichELISA, IndirectSandwichELISA, the options will be automatically set to Model[Sample,"ELISA Blocker Blocking Buffer"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
PrimaryAntibodyStorageCondition
The condition under which the unused portion of PrimaryAntibody stock sample should be stored.
Default Value: Refrigerator
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal.
Programmatic Pattern: SampleStorageTypeP | Disposal
Index Matches to: experiment samples
SecondaryAntibody
The antibody that binds to the primary antibody.
Default Calculation: If Method is IndirectELISA, IndirectSandwichELISA, or IndirectCompetitiveELISA, the option is automatically set to a stocked secondary antibody for the primary antibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
SecondaryAntibodyDilutionFactor
The dilution ratio of SecondaryAntibody. SecondaryAntibody is always diluted in the SecondryAntibodyDiluent. Either SecondaryAntibodyDilutionFactor or SecondaryAntibodyVolume should be provided but not both.
Default Calculation: For IndirectELISA, IndirectSandwichELISA, IndirectCompetitiveELISA, automatically set to 0.001 (1:1,000).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: experiment samples
SecondaryAntibodyVolume
The volume of SecondaryAntibody added into the corresponding well of the assay plate. SecondaryAntibodyVolume is used as an alternative to SecondaryAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: experiment samples
SecondaryAntibodyDiluent
The buffer used to dilute SecondaryAntibody.
Default Calculation: If Method is set to IndirectELISA, IndirectSandwichELISA, IndirectCompetitiveELISA, the option will automatically set to Model[Sample,"ELISA Blocker Blocking Buffer"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
SecondaryAntibodyStorageCondition
The condition under which the unused portion of Secondary Antibody stock sample should be stored.
Default Calculation: If method is IndirectELISA, IndirectSandwichELISA, or IndirectCompetitiveELISA, the option is automatically set to Refrigerator.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
Sample Antibody Complex Incubation
SampleAntibodyComplexIncubation
Indicates if the pre-mixed samples and antibodies should be incubated before loaded into the assay plate. The plate used for sample-antibody mixing will be discarded after the experiment.
Default Calculation: Automatically set to True if Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, and FastELISA.
Pattern Description: {True, False}
Programmatic Pattern: BooleanP | Automatic
SampleAntibodyComplexIncubationTime
The duration of sample-antibody complex incubation (If needed). In DirectCompetitiveELISA and IndirectCompetitiveELISA, PrimaryAntibody is incubated with the sample. In FastELISA, PrimaryAntibody and CaptureAntibody is incubated with the sample. If Null, the prepared sample-antibody complex will be kept at 4 degree Celsius till ready to use.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 2 Hours.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 24 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, 24*Hour] | Automatic) | Null
SampleAntibodyComplexIncubationTemperature
The temperature at which sample mixed with Antibodies are incubated. In DirectCompetitiveELISA and IndirectCompetitiveELISA, PrimaryAntibody is incubated with the sample. In FastELISA, PrimaryAntibody and CaptureAntibody are incubated with the sample. If Null, the prepared sample-antibody complex will be used directly.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to Ambient.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[4*Celsius, 50*Celsius]) | Automatic) | Null
Coating
Coating
Indicates if Coating is required. Coating is a procedure to non-specifically adsorb protein molecules to the surface of wells of the assay plate.
Pattern Description: {True, False}
Programmatic Pattern: BooleanP
SampleCoatingVolume
The amount of Sample that is aliquoted into the assay plate, in order for the Sample to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectELISA or IndirectELISA, CoatingVolume is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
CoatingAntibodyCoatingVolume
The amount of diluted CoatingAntibody that is aliquoted into the assay plate, in order for the CoatingAntibody to be adsorbed to the surface of the well.
Default Calculation: If Method is FastELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 300*Microliter] | Automatic) | Null
Index Matches to: experiment samples
ReferenceAntigenCoatingVolume
The amount of diluted ReferenceAntigen that is aliquoted into the assay plate, in order for the ReferenceAntigen to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectCompetitiveELISA or IndirectCompetitiveELISA, CoatingVolume is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyCoatingVolume
The amount of diluted CaptureAntibody that is aliquoted into the assay plate, in order for the CaptureAntibody to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectSandwichELISA or IndirectSandwichELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
CoatingTemperature
The temperature at which the Coating Solution is kept in the assay plate, in order for the coating molecules to be adsorbed to the surface of the well.
Default Calculation: If Coating is set to True, the option is automatically set to 4 degree Celsius.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[4*Celsius, 50*Celsius]) | Automatic) | Null
CoatingTime
The duration when the Coating Solution is kept in the assay plate, in order for the coating molecules to be adsorbed to the surface of the well.
Default Calculation: If Coating is set to True, the option is automatically set to 16 Hours.
Pattern Description: Greater than or equal to 0 hours and less than or equal to 20 hours or Null.
Programmatic Pattern: (RangeP[0*Hour, 20*Hour] | Automatic) | Null
CoatingWashVolume
The volume of WashBuffer added to rinse off unbound molecule. When Coating is False but Blocking is True, the pre-coated plate must be washed at least once to ensure no liquid is left in the plate.
Default Calculation: If Coating is set to True, or Coating is False but Blocking is True, the option is automatically set to 250 Microliters.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 300*Microliter] | Automatic) | Null
CoatingNumberOfWashes
The number of washes performed after coating. When Coating is False but Blocking is True, the pre-coated plate must be washed at least once to ensure no liquid is left in the plate.
Default Calculation: If Coating is set to True,or Coating is False but Blocking is True, the option is automatically set to 3.
Pattern Description: Greater than 0 in increments of 1 or Null.
Programmatic Pattern: (GreaterP[0, 1] | Automatic) | Null
Blocking
Blocking
Indicates if a protein solution should be incubated with the assay plate to prevent non-specific binding of molecules to the assay plate.
Pattern Description: {True, False}
Programmatic Pattern: BooleanP
BlockingBuffer
The protein-containing solution used to prevent non-specific binding of antigen or antibody to the surface of the assay plate.
Default Calculation: If Blocking is True, automatically set to Model[Sample,"ELISA Blocker Blocking Buffer"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlockingVolume
The amount of BlockingBuffer that is aliquoted into the appropriate wells of the assay plate, in order to prevent non-specific binding of molecules to the assay plate.
Default Calculation: If Blocking is True, the option is automatically set to 100 Microliters.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 300*Microliter] | Automatic) | Null
Index Matches to: experiment samples
BlockingTime
The duration when the BlockingBuffer is kept with the assay plate, in order to prevent non-specific binding of molecules to the assay plate.
Default Calculation: If Blocking is True, the option is automatically set to 1 Hour.
Pattern Description: Greater than or equal to 0 hours and less than or equal to 20 hours or Null.
Programmatic Pattern: (RangeP[0*Hour, 20*Hour] | Automatic) | Null
BlockingTemperature
The duration of time when the BlockingBuffer is kept with the assay plate, in order to prevent non-specific binding of molecules to the assay plate.
Default Calculation: If Blocking is True, the option is automatically set to 25 degree Celsius.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 50*Celsius] | Automatic) | Null
BlockingMixRate
The speed at which the plate is shaken (orbitally, at a radius of 2 mm) during Blocking incubation. Mixing is not recommended when incubation volume is high, in which case the option should be set to Null.
Pattern Description: Greater than or equal to 40 revolutions per minute and less than or equal to 1000 revolutions per minute or Null.
Programmatic Pattern: RangeP[40*RPM, 1000*RPM] | Null
BlockingWashVolume
The volume of WashBuffer added after Blocking, in order to rinse off the unbound molecules from the surface of the wells. If Coating and Blocking are both False, at least one blocking wash is required to ensure no liquid is remaining in the assay plate.
Default Calculation: If Blocking is True, or Coating and Blocking are both False, the option is automatically set to 250 Microliters.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 300*Microliter] | Automatic) | Null
BlockingNumberOfWashes
The number of washes performed after Blocking, in order to rinse off the unbound molecules from the surface of the wells.
Default Calculation: If Blocking is True, or Coating and Blocking are both False, the option is automatically set to 3.
Pattern Description: Greater than 0 in increments of 1 or Null.
Programmatic Pattern: (GreaterP[0, 1] | Automatic) | Null
Immunosorbent Step
SampleAntibodyComplexImmunosorbentVolume
The volume of the sample-antibody complex to be loaded on each well of the ELISAPlate. In DirectCompetitiveELISA and IndirectCompetitiveELISA, this step enables the free primary antibody to bind to the ReferenceAntigen coated on the plate. In FastELISA, this step enables the PrimaryAntibody-TargetAntigen-CaptureAntibody complex to bind to the CoatingAntibody on the plate.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SampleAntibodyComplexImmunosorbentTime
The duration of sample-antibody complex incubation.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 2 Hour.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 24 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, 24*Hour] | Automatic) | Null
SampleAntibodyComplexImmunosorbentTemperature
The temperature of the sample-antibody complex incubation.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 25 Celsius.
Pattern Description: Ambient or greater than or equal to 4 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[4*Celsius, 50*Celsius]) | Automatic) | Null
SampleAntibodyComplexImmunosorbentMixRate
The speed at which the plate is shaken (orbitally, at a radius of 2 mm) during SampleAntibody mixture incubation in the assay plate. Mixing is not recommended when incubation volume is higher than 200 Microliters, in which case the options should be set to Null.
Pattern Description: Greater than or equal to 40 revolutions per minute and less than or equal to 1000 revolutions per minute or Null.
Programmatic Pattern: RangeP[40*RPM, 1000*RPM] | Null
SampleAntibodyComplexImmunosorbentWashVolume
The volume of WashBuffer added to rinse off the unbound primary antibody after sample-antibody complex incubation.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 250 Microliter.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 300*Microliter] | Automatic) | Null
SampleAntibodyComplexImmunosorbentNumberOfWashes
The number of rinses performed after sample-antibody complex incubation.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 4.
Pattern Description: Greater than 0 in increments of 1 or Null.
Programmatic Pattern: (GreaterP[0, 1] | Automatic) | Null
SampleImmunosorbentVolume
The volume of the Sample to be loaded on the ELISAPlate for the target antigen to bind to the capture antibody in DirectSandwichELISA and IndirectSandwichELISA.
Default Calculation: If the Method is set to DirectSandwichELISA and IndirectSandwichELISA, the option is automatically set to 100 Microliters.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SampleImmunosorbentTime
The duration of Sample incubation.
Default Calculation: If the Method is set to DirectSandwichELISA and IndirectSandwichELISA, the option is automatically set to 2 Hour.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 24 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, 24*Hour] | Automatic) | Null
SampleImmunosorbentTemperature
The temperature of the Sample incubation.
Default Calculation: If the Method is set to DirectSandwichELISA and IndirectSandwichELISA, the option is automatically set to 25 Celsius.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 50*Celsius] | Automatic) | Null
SampleImmunosorbentMixRate
The speed at which the plate is shaken (orbitally, at a radius of 2 mm) during Sample incubation. Mixing is not recommended when incubation volume is higher than 200 Microliters, in which case the options should be set to Null.
Pattern Description: Greater than or equal to 40 revolutions per minute and less than or equal to 1000 revolutions per minute or Null.
Programmatic Pattern: RangeP[40*RPM, 1000*RPM] | Null
SampleImmunosorbentWashVolume
The volume of WashBuffer added to rinse off the unbound Sample after Sample incubation.
Default Calculation: If the Method is set to DirectSandwichELISA and IndirectSandwichELISA, the option is automatically set to 250 Microliters.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 300*Microliter] | Automatic) | Null
SampleImmunosorbentNumberOfWashes
The number of rinses performed after Sample incubation.
Default Calculation: If the Method is set to DirectSandwichELISA and IndirectSandwichELISA, the option is automatically set to 4.
Pattern Description: Greater than 0 in increments of 1 or Null.
Programmatic Pattern: (GreaterP[0, 1] | Automatic) | Null
PrimaryAntibodyImmunosorbentVolume
The volume of the PrimaryAntibody to be loaded on the ELISA Plate for immunosorbent step.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
PrimaryAntibodyImmunosorbentTime
The duration of PrimaryAntibody incubation.
Default Calculation: If the Method is set to DirectELISA, IndirectELISA, DirectSandwichELISA, or IndirectSandwichELISA, the option is automatically set to 2 Hour.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 24 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, 24*Hour] | Automatic) | Null
PrimaryAntibodyImmunosorbentTemperature
The temperature of the PrimaryAntibody incubation.
Default Calculation: If the Method is set to DirectELISA, IndirectELISA, DirectSandwichELISA, or IndirectSandwichELISA, the option is automatically set to 25 Celsius.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 50*Celsius] | Automatic) | Null
PrimaryAntibodyImmunosorbentMixRate
The speed at which the plate is shaken (orbitally, at a radius of 2 mm) during PrimaryAntibody incubation. Mixing is not recommended when incubation volume is higher than 200 Microliters, in which case the options should be set to Null.
Pattern Description: Greater than or equal to 40 revolutions per minute and less than or equal to 1000 revolutions per minute or Null.
Programmatic Pattern: RangeP[40*RPM, 1000*RPM] | Null
PrimaryAntibodyImmunosorbentWashVolume
The volume of WashBuffer added to rinse off the unbound primary antibody after PrimaryAntibody incubation.
Default Calculation: If the Method is set to DirectELISA, IndirectELISA, DirectSandwichELISA, or IndirectSandwichELISA, the option is automatically set to 250 Microliters.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 300*Microliter] | Automatic) | Null
PrimaryAntibodyImmunosorbentNumberOfWashes
The number of rinses performed after PrimaryAntibody incubation.
Default Calculation: If the Method is set to DirectELISA, IndirectELISA, DirectSandwichELISA, or IndirectSandwichELISA, the option is automatically set to 4.
Pattern Description: Greater than 0 in increments of 1 or Null.
Programmatic Pattern: (GreaterP[0, 1] | Automatic) | Null
SecondaryAntibodyImmunosorbentVolume
The volume of the Secondary Antibody to be loaded on the ELISAPlate for the immunosorbent step.
Default Calculation: If the Method is set to IndirectELISA, IndirectSandwichELISA, and IndirectCompetitiveELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SecondaryAntibodyImmunosorbentTime
The duration of Secondary Antibody incubation.
Default Calculation: If the Method is set to IndirectELISA, IndirectSandwichELISA, and IndirectCompetitiveELISA, the option is automatically set to 1 Hour.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 24 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, 24*Hour] | Automatic) | Null
SecondaryAntibodyImmunosorbentTemperature
The temperature of the Secondary Antibody incubation.
Default Calculation: If the Method is set to IndirectELISA, IndirectSandwichELISA, and IndirectCompetitiveELISA, the option is automatically set to 25 Celsius.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 50*Celsius] | Automatic) | Null
SecondaryAntibodyImmunosorbentMixRate
The speed at which the plate is shaken (orbitally, at a radius of 2 mm) during Secondary Antibody incubation. Mixing is not recommended when incubation volume is higher than 200 Microliters, in which case the options should be set to Null.
Pattern Description: Greater than or equal to 40 revolutions per minute and less than or equal to 1000 revolutions per minute or Null.
Programmatic Pattern: RangeP[40*RPM, 1000*RPM] | Null
SecondaryAntibodyImmunosorbentWashVolume
The volume of WashBuffer added to rinse off the unbound Secondary antibody after SecondaryAntibody incubation.
Default Calculation: If the Method is set to IndirectELISA, IndirectSandwichELISA, and IndirectCompetitiveELISA, the option is automatically set to 250 Microliters.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 300*Microliter] | Automatic) | Null
SecondaryAntibodyImmunosorbentNumberOfWashes
The number of rinses performed after SecondaryAntibody incubation.
Default Calculation: If the Method is set to IndirectELISA, IndirectSandwichELISA, and IndirectCompetitiveELISA, the option is automatically set to 4.
Pattern Description: Greater than 0 in increments of 1 or Null.
Programmatic Pattern: (GreaterP[0, 1] | Automatic) | Null
Detection
SubstrateSolution
Defines the substrate solution to the enzyme conjugated to the antibody.
Default Calculation: If enzyme is Horseredish Peroxidase, the option will be automatically set to Model[Sample,"ELISA TMB Stabilized Chromogen"]. If enzyme is Alkaline Phosphatase, then the option is automatically set to Model[Sample,"AP Substrate PNPP Solution"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic
Index Matches to: experiment samples
StopSolution
The reagent that is used to stop the reaction between the enzyme and its substrate.
Default Calculation: If enzyme is Horseradish Peroxidase, then the option is automatically set to Model[Sample,"ELISA HRP-TMB Stop Solution"]. If enzyme is Alkaline Phosphatase, then the option is automatically set to Model[Sample,"AP Substrate Stop Solution"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
SubstrateSolutionVolume
The volume of substrate to be added to the corresponding well.
Default Calculation: If SubstrateSolution is populated, then the option is automatically set to 100ul.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Automatic
Index Matches to: experiment samples
StopSolutionVolume
The volume of StopSolution to be added to the corresponding well.
Default Calculation: If StopSolution is selected, the StopSolutionVolume is automatically set to 100ul.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SubstrateIncubationTime
The time during which the colorimetric reaction occurs.
Default Value: 30 minutes
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 24 hours.
Programmatic Pattern: RangeP[0*Minute, 24*Hour]
SubstrateIncubationTemperature
The temperature of the Substrate incubation, in order for the detection reaction to take place where the antibody-conjugated enzyme (such as Horseradish Peroxidase or Alkaline Phosphatase) catalyzes the colorimetric reaction.
Default Value: 25 degrees Celsius
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 50 degrees Celsius.
Programmatic Pattern: RangeP[25*Celsius, 50*Celsius]
SubstrateIncubationMixRate
The speed at which the plate is shaken (orbitally, at a radius of 2 mm) during Substrate incubation. Mixing is not recommended when incubation volume is higher than 200 Microliters, in which case the options should be set to Null.
Pattern Description: Greater than or equal to 40 revolutions per minute and less than or equal to 1000 revolutions per minute or Null.
Programmatic Pattern: RangeP[40*RPM, 1000*RPM] | Null
AbsorbanceWavelength
The wavelength used to detect the absorbance of light by the product of the detection reaction.
Default Value: 450 nanometers
Pattern Description: {405 nanometers, 450 nanometers, 492 nanometers, 620 nanometers} or list of one or more {405 nanometers, 450 nanometers, 492 nanometers, 620 nanometers} entries.
Programmatic Pattern: (405*Nanometer | 450*Nanometer | 492*Nanometer | 620*Nanometer) | {(405*Nanometer | 450*Nanometer | 492*Nanometer | 620*Nanometer)..}
PrereadBeforeStop
Indicate if colorimetric reactions between the enzyme and its substrate will be checked by the plate reader before the stopping reagent was added to terminate the reaction.
Default Calculation: Is set automatically to True if either or both of PrereadTimepoints and PrereadAbsorbanceWavelength are specified. Otherwise is set to False.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
PrereadTimepoints
The list of time points when absorbance intensitied at each PreparedAbsorbanceWavelength during the Preread process (before the colorimetric reaction was terminated).
Default Calculation: If PrereadBeforeStop is True, this option is set to one single value: the half of the SubstrateIncubationTime. Otherwise is set to Null.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 72 hours or list of one or more greater than or equal to 0 minutes and less than or equal to 72 hours entries or Null.
Programmatic Pattern: ((RangeP[0*Minute, $MaxExperimentTime] | {RangeP[0*Minute, $MaxExperimentTime]..}) | Automatic) | Null
PrereadAbsorbanceWavelength
The wavelength used to detect the absorbance of light during the Preread process (before the colorimetric reaction was terminated).
Default Calculation: If PrereadBeforeStop is True, this option is set to {450 Nanometer}. Otherwise is set to Null.
Pattern Description: {405 nanometers, 450 nanometers, 492 nanometers, 620 nanometers} or list of one or more {405 nanometers, 450 nanometers, 492 nanometers, 620 nanometers} entries or Null.
Programmatic Pattern: (((405*Nanometer | 450*Nanometer | 492*Nanometer | 620*Nanometer) | {(405*Nanometer | 450*Nanometer | 492*Nanometer | 620*Nanometer)..}) | Automatic) | Null
SignalCorrection
The absorbance reading that is used to eliminate the interference of background absorbance (such as from ELISAPlate material and dust). If True, a reading at 620 nm is read at the same time of the AbsorbanceWavelength. The correction is done by subtracting the reading at 620nm from that at the AbsorbanceWavelength.
Pattern Description: {True, False} or Null.
Programmatic Pattern: BooleanP | Null
Standard
Standard
A sample containing known amount of TargetAntigen molecule. Standard is used for the quantification of Standard analyte.
Default Calculation: If TargetAntigen is specified, standard will be automatically set to the DefaultSampleModel of the target antigen.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardTargetAntigen
The Analyte molecule(e.g., peptide, protein, and hormone) detected and quantified in the Standard samples by Antibodies in the ELISA experiment. This option is used to automatically set sample Antibodies and the corresponding experiment conditions of Standards and Blanks.
Default Calculation: Automatically set to the Model[Molecule] in the Analyte field of the sample.
Pattern Description: An object of type or subtype Model[Molecule] or Null.
Programmatic Pattern: (ObjectP[{Model[Molecule]}] | Automatic) | Null
Index Matches to: Standard
StandardSerialDilutionCurve
The collection of serial dilutions that will be performed on sample. StandardLoadingVolume of each dilution will be transferred to the ELISAPlate. For Serial Dilution Volumes, the Transfer Volume is taken out of the sample and added to a second well with the Diluent Volume of the Diluent. It is mixed, then the Transfer Volume is taken out of that well to be added to a third well. This is repeated to make Number Of Dilutions diluted samples. For example, if a 100 ug/ ml sample with a Transfer Volume of 20 Microliters, a Diluent Volume of 60 Microliters and a Number of Dilutions of 3 is used, it will create a DilutionCurve of 25 ug/ ml, 6.25 ug/ ml, and 1.5625 ug/ ml with each dilution having a volume of 60 Microliters. For Serial Dilution Factors, the sample will be diluted by the dilution factor at each transfer step. IMPORTANT: because the dilution curve does not intrinsically include the original sample, if the original standard solution is to be included in the dilution curve, the first diluting factor should be set to 1 or Diluent Volume should be 0 Microliters. During experiment, an 5% extra volume than specified is going to be prepared in order to offset pipetting errors. Therefore, the total volume for each well specified in this option should be equal or greater than the volume needed for the experiment. Because the dilution of each assay (well) is prepared independently, the NumberOfReplications should not be considered when determining the Standard Volume. The plate used for sample dilutions will be discarded after the experiment. Note: if spike or antibodies are to be mixed with samples, their volumes are counted towards diluent volume.
Default Calculation: Automatically set to Null if the StandardDilutionCurve is specified. In all other cases it is automatically set to a Standard Volume of 100 Microliters and a Constant Dilution Factor of 0.5. The Number Of Dilutions is automatically set to 6.
Pattern Description: Serial Dilution Factor or Serial Dilution Volumes or Null.
Programmatic Pattern: (({GreaterEqualP[0*Microliter], {RangeP[0, 1], GreaterP[0, 1]} | {RangeP[0, 1]..}} | {RangeP[0*Microliter, 1000*Microliter], RangeP[40*Microliter, 1000*Microliter], GreaterP[0, 1]}) | Automatic) | Null
Index Matches to: Standard
StandardDilutionCurve
The collection of dilutions that will be performed on each sample. For Fixed Dilution Volume Dilution Curves, the Standard Amount is the volume of the sample that will be mixed the Diluent Volume of the Diluent to create a desired concentration. The Standard Volume is the TOTAL volume of the sample that will created after being diluted by the Dilution Factor. For example, a 1 ug/ ml sample with a dilution factor of 0.7 will be diluted to a concentration 0.7 ug/ ml. IMPORTANT: because the dilution curve does not intrinsically include the original sample, if the original standard solution is to be included in the dilution curve, the first diluting factor should be set to 1 or Diluent Volume must be 0 Microliter. During experiment, an 5% extra volume than specified is going to be prepared in order to offset pipetting errors. Therefore, the total volume for each well specified in this option should be equal or greater than the volume needed for the experiment. Because the dilution of each assay (well) is prepared independently, the NumberOfReplications should not be considered when determining the Standard Volume. The plate used for sample dilutions will be discarded after the experiment. Note: if spike or antibodies are to be mixed with samples, their volumes are counted towards diluent volume.
Default Calculation: Automatically set to Null if the SerialDilutionCurve is specified. If SerialDilutionCurve is set to Null, then automatically set Standard Assay Volume to 100ul, dilution factor to 0.5, 0,25, 0.125, 0.0625, 0.03125, 0.015625.
Pattern Description: Fixed Dilution Factor or Fixed Dilution Volume or Null.
Programmatic Pattern: (({{RangeP[0*Microliter, 1000*Microliter], RangeP[0*Microliter, 1000*Microliter]}..} | {{RangeP[40*Microliter, 1500*Microliter], RangeP[0, 1]}..}) | Automatic) | Null
Index Matches to: Standard
StandardDiluent
The buffer used to perform multiple dilutions on Standards.
Default Calculation: If StandardDilutionCurve and StandardSerialDilutionCurve are not both Null, in the case when Method is set to DirectELISA and IndirectELISA, the option is automatically set to Model[Sample, StockSolution, "1x Carbonate-Bicarbonate Buffer pH10"]; in all other cases, the option is automatically set to Model[Sample,"ELISA Blocker Blocking Buffer"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
StandardStorageCondition
The condition under which the unused portion of Standard stock sample should be stored.
Default Calculation: If Standard is Specified, the option is automatically set to Freezer.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: Standard
StandardCoatingAntibody
The sample containing the antibody that is used for coating in FastELISA.
Default Calculation: If Method is FastELISA and Standard is not Null, automatically set to an antibody against a tag which is conjugated to a the StandardCaptureAntibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardCoatingAntibodyDilutionFactor
The dilution ratio of StandardCoatingAntibody. StandardCoatingAntibody is diluted with CoatingAntibodyDiluent. Either StandardCoatingAntibodyDilutionFactor or StandardCoatingAntibodyVolume should be provided but not both.
Default Calculation: If Method is FastELISA and Standard is not Null, automatically set to 0.001 (1:1,000)
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: Standard
StandardCoatingAntibodyVolume
The volume of undiluted StandardCoatingAntibody added into the corresponding well of the assay plate. StandardCoatingAntibody is diluted with CoatingAntibodyDiluent. StandardCoatingAntibodyVolume is used as an alternative to StandardCoatingAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: Standard
StandardCaptureAntibody
The sample containing the antibody that is used to pull down the antigen from sample solution to the plate in DirectSandwichELISA, IndirectSandwichELISA, and FastELISA.
Default Calculation: Automatically set to Null is Standard is Null. If Method is FastELISA, automatically set to an antibody containing an affinity tag and against the TargetAntigen. If Method is DirectSandwichELISA or IndirectSandwichELISA, automatically set to an un-tagged antibody against TargetAntigen.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyDilutionFactor
The dilution ratio of StandardCaptureAntibody. For DirectSandwichELISA and IndirectSandwichELISA, StandardCaptureAntibody is diluted with CaptureAntibodyDiluent. For FastELISA, StandardCaptureAntibody is diluted in the corresponding standard. Either StandardCaptureAntibodyDilutionFactor or StandardCaptureAntibodyVolume should be provided but not both.
Default Calculation: When Standard is not Null, if Method is DirectSandwichELISA or IndirectSandwichELISA, automatically set to 0.001 (1:1,000). If Method is FastELISA, automatically set to 0.01 (1:100).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyVolume
The volume of undiluted CaptureAntibody added into the corresponding well of the assay plate. StandardCaptureAntibodyVolume is used as an alternative to StandardCaptureAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: Standard
StandardReferenceAntigen
The sample containing the antigen that is used in DirectCompetitiveELISA or IndirectCompetitiveELISA. The StandardReferenceAntigen competes with sample antigen for the binding of the StandardPrimaryAntibody. Reference antigen is sometimes also referred to as inhibitor antigen.
Default Calculation: Automatically set to a sample containing known amount of TargetAntigen when Method is set to DirectCompetitiveELISA or IndirectCompetitiveELISA and Standard is not Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardReferenceAntigenDilutionFactor
The dilution ratio of StandardReferenceAntigen. For DirectCompetitiveELISA and IndirectCompetitiveELISA, the StandardReferenceAntigenStandard is diluted in ReferenceAntigenDiluent. Either StandardReferenceAntigenDilutionFactor or StandardReferenceAntigenVolume should be provided but not both.
Default Calculation: Automatically set to 0.001 (1:1,000) if Method is DirectCompetitiveELISA or IndirectCompetitiveELISA.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: Standard
StandardReferenceAntigenVolume
The volume of ReferenceAntigen added into the corresponding well of the assay plate. StandardReferenceAntigenVolume is used as an alternative to StandardReferenceAntigenDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: Standard
StandardPrimaryAntibody
The antibody that directly binds to the analyte.
Default Calculation: If Standard is not Null, the option will be automatically set to an antibody against the TargetAntigen.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardPrimaryAntibodyDilutionFactor
The dilution ratio of StandardPrimaryAntibody. For DirectELISA, IndirectELISA, DirectSandwichELISA, and IndirectSandwichELISA, the antibody is diluted with PrimaryAntibodyDiluent. For DirectCompetitiveELISA, IndirectCompetitiveELISA, and FastELISA, the antibody is diluted in the corresponding sample. Either StandardPrimaryAntibodyDilutionFactor or StandardPrimaryAntibodyVolume should be provided but not both.
Default Calculation: When Standard is not Null, if used for DirectELISA, IndirectELISA, DirectSandwichELISA, IndirectSandwichELISA, automatically set to 0.001 (1:1,000). If used for DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, automatically set to 0.01 (1:100).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: Standard
StandardPrimaryAntibodyVolume
The volume of PrimaryAntibody added into the corresponding well of the assay plate. StandardPrimaryAntibodyVolume is used as an alternative to StandardPrimaryAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: Standard
StandardSecondaryAntibody
The antibody that binds to the primary antibody.
Default Calculation: If Method is IndirectELISA, IndirectSandwichELISA, or IndirectCompetitiveELISA, the option is automatically set to a stocked secondary antibody for the primary antibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardSecondaryAntibodyDilutionFactor
The dilution ratio of StandardSecondaryAntibody. StandardSecondaryAntibody is always diluted in the SecondaryAntibodyDiluent. Either StandardSecondaryAntibodyDilutionFactor or StandardSecondaryAntibodyVolume should be provided but not both.
Default Calculation: If Standard is not Null, for IndirectELISA, IndirectSandwichELISA, IndirectCompetitiveELISA, automatically set to 0.001 (1:1,000).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
Index Matches to: Standard
StandardSecondaryAntibodyVolume
The volume of SecondaryAntibody added into the corresponding well of the assay plate. StandardSecondaryAntibodyVolume is used as an alternative to StandardSecondaryAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
Index Matches to: Standard
StandardCoatingVolume
The amount of Standard that is aliquoted into the ELISAPlate, in order for the Standard to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectELISA or IndirectELISA, StandardCoatingVolume is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardReferenceAntigenCoatingVolume
The amount of StandardReferenceAntigen that is aliquoted into the assay plate, in order for the StandardReferenceAntigen to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectCompetitiveELISA or IndirectCompetitiveELISA, CoatingVolume is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardCoatingAntibodyCoatingVolume
The amount of diluted StandardCoatingAntibody that is aliquoted into the ELISAPlate, in order for the StandardCoatingAntibody to be adsorbed to the surface of the well.
Default Calculation: If Method is FastELISA, the option is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyCoatingVolume
The amount of diluted StandardCaptureAntibody that is aliquoted into the ELISAPlate, in order for the StandardCaptureAntibody to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectSandwichELISA or IndirectSandwichELISA, the option is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardBlockingVolume
The amount of BlockingBuffer that is aliquoted into the appropriate wells of the ELISAPlate.
Default Calculation: If Blocking is True, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 300*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardAntibodyComplexImmunosorbentVolume
The volume of the standard-antibody complex to be loaded on the ELISAPlate. In DirectCompetitiveELISA and IndirectCompetitiveELISA, this step enables the free StandardPrimaryAntibody to bind to the StandardReferenceAntigen coated on the plate. In FastELISA, this step enables the PrimaryAntibody-TargetAntigen-CaptureAntibody complex to bind to the CoatingAntibody on the plate.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardImmunosorbentVolume
The volume of the Standard to be loaded on the ELISAPlate for the target antigen to bind to the capture antibody in DirectSandwichELISA and IndirectSandwichELISA.
Default Calculation: If the Method is set to DirectSandwichELISA and IndirectSandwichELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardPrimaryAntibodyImmunosorbentVolume
The volume of the StandarPrimaryAntibody to be loaded on the ELISAPlate for Immunosorbent assay.
Default Calculation: If the Method is set to DirectELISA, IndirectELISA, DirectSandwichELISA, or IndirectSandwichELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardSecondaryAntibodyImmunosorbentVolume
The volume of the StandardSecondaryAntibody to be loaded on the ELISAPlate for immunosorbent step.
Default Calculation: If the Method is set to IndirectELISA, IndirectSandwichELISA, and IndirectCompetitiveELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardSubstrateSolution
Defines the substrate solution for the standard solution.
Default Calculation: If enzyme is Horseredish Peroxidase, the option will be automatically set to Model[Sample,"ELISA TMB Stabilized Chromogen"]. If enzyme is Alkaline Phosphatase, then the option is automatically set to Model[Sample,"AP Substrate PNPP Solution"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardStopSolution
The reagent that is used to stop the reaction between the enzyme and its substrate.
Default Calculation: If enzyme is Horseradish Peroxidase, then the option is automatically set to Model[Sample,"ELISA HRP-TMB Stop Solution"]. If enzyme is Alkaline Phosphatase, then the option is automatically set to Model[Sample,"AP Substrate Stop Solution"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardSubstrateSolutionVolume
The volume of StandardSubstrateSolution to be added to the corresponding well.
Default Calculation: If StandardSubstrateSolution is populated and Standard is not Null, then the option is automatically set to 100ul.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardStopSolutionVolume
The volume of StopSolution to be added to the corresponding well.
Default Calculation: If Model[Sample,"ELISA TMB Stabilized Chromogen"] or Model[Sample,"ELISA HRP-TMB Stop Solution"] is selected for StandardSubstrateSolution, the StopSolutionVolume is automatically set to 100ul.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Index Matches to: Standard
Blank
Blank
A sample containing no TargetAntigen, used as a baseline or negative control for the ELISA.
Default Calculation: If In DirectELISA and IndirectELISA, the option will be automatically set to Model[Sample, StockSolution, "1x Carbonate-Bicarbonate Buffer pH10"]. Otherwise, the option will be automatically set to Model[Sample, "ELISA Blocker Blocking Buffer"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankTargetAntigen
The Analyte molecule(e.g., peptide, protein, and hormone) detected and quantified in the blanks by Antibodies in the ELISA experiment. This option is used to automatically set Antibodies and the corresponding experiment conditions.
Default Calculation: Automatically set to the Model[Molecule] in the Analyte field of the sample.
Pattern Description: An object of type or subtype Model[Molecule] or Null.
Programmatic Pattern: (ObjectP[{Model[Molecule]}] | Automatic) | Null
BlankStorageCondition
The condition under which the unused portion of Blank should be stored.
Default Calculation: Automatically set to Refrigerator if Blank is not Null.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
BlankCoatingAntibody
The sample containing the antibody that is used for coating in FastELISA.
Default Calculation: If Method is FastELISA and Blank is not Null, automatically set to an antibody against a tag which is conjugated to a the StandardCaptureAntibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankCoatingAntibodyDilutionFactor
The dilution ratio of BlankCoatingAntibody. BlankCoatingAntibody is diluted with CoatingAntibodyDiluent. Either BlankCoatingAntibodyDilutionFactor or BlankCoatingAntibodyVolume should be provided but not both.
Default Calculation: If Method is FastELISA and Blank is not Null, automatically set to 0.001 (1:1,000)
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
BlankCoatingAntibodyVolume
The volume of BlankCoatingAntibody added into the corresponding well of the assay plate. BlankCoatingAntibody is diluted with CoatingAntibodyDiluent. BlankCoatingAntibodyVolume is used as an alternative to BlankCoatingAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
BlankCaptureAntibody
The sample containing the antibody that is used to pull down the antigen from sample solution to the plate in DirectSandwichELISA, IndirectSandwichELISA, and FastELISA.
Default Calculation: Automatically set to Null is Blank is Null. If Method is FastELISA, automatically set to an antibody containing an affinity tag and against the TargetAntigen. If Method is DirectSandwichELISA or IndirectSandwichELISA, automatically set to an un-tagged antibody against TargetAntigen.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankCaptureAntibodyDilutionFactor
The dilution ratio of BlankCaptureAntibody. For DirectSandwichELISA and IndirectSandwichELISA, BlankCaptureAntibody is diluted with CaptureAntibodyDiluent. For FastELISA, BlankCaptureAntibody is diluted in the corresponding sample. Either BlankCaptureAntibodyDilutionFactor or BlankCaptureAntibodyVolume should be provided but not both.
Default Calculation: If Method is DirectSandwichELISA or IndirectSandwichELISA, automatically set to 0.001 (1:1,000). If Method is FastELISA, automatically set to 0.01 (1:100).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
BlankCaptureAntibodyVolume
The volume of CaptureAntibody added into the corresponding well of the assay plate. BlankCaptureAntibodyVolume is used as an alternative to BlankCaptureAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
BlankReferenceAntigen
The Sample containing the antigen that is used in DirectCompetitiveELISA or IndirectCompetitiveELISA. The BlankReferenceAntigen competes with sample antigen for the binding of the BlankPrimaryAntibody. Reference antigen is sometimes also referred to as inhibitor antigen.
Default Calculation: Automatically set to a sample containing known amount of TargetAntigen when Method is set to DirectCompetitiveELISA or IndirectCompetitiveELISA and Blank is not Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankReferenceAntigenDilutionFactor
The dilution ratio of BlankReferenceAntigen. For DirectCompetitiveELISA and IndirectCompetitiveELISA, the BlankReferenceAntigenBlank is diluted in ReferenceAntigenDiluent. Either BlankReferenceAntigenDilutionFactor or BlankReferenceAntigenVolume should be provided but not both.
Default Calculation: Automatically set to 0.001 (1:1,000) if Method is DirectCompetitiveELISA or IndirectCompetitiveELISA.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
BlankReferenceAntigenVolume
The volume of ReferenceAntigen added into the corresponding well of the assay plate. BlankReferenceAntigenVolume is used as an alternative to BlankReferenceAntigenDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
BlankPrimaryAntibody
The antibody that directly binds with the analyte.
Default Calculation: The option will be automatically set to an antibody against the BlankTargetAntigen if Blank is not Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankPrimaryAntibodyDilutionFactor
The dilution ratio of BlankPrimaryAntibody. For DirectELISA, IndirectELISA, DirectSandwichELISA, and IndirectSandwichELISA, the antibody is diluted with PrimaryAntibodyDiluent. For DirectCompetitiveELISA, IndirectCompetitiveELISA, and FastELISA, the antibody is diluted in the corresponding sample. Either BlankPrimaryAntibodyDilutionFactor or BlankPrimaryAntibodyVolume should be provided but not both.
Default Calculation: When Blank is not Null, if used for DirectELISA, IndirectELISA, DirectSandwichELISA, IndirectSandwichELISA, automatically set to 0.001 (1:1,000). If used for DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, automatically set to 0.01 (1:100).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
BlankPrimaryAntibodyVolume
The volume of PrimaryAntibody added into the corresponding well of the assay plate. BlankPrimaryAntibodyVolume is used as an alternative to BlankPrimaryAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
BlankSecondaryAntibody
The antibody that binds to the primary antibody.
Default Calculation: If Method is IndirectELISA, IndirectSandwichELISA, or IndirectCompetitiveELISA and Blank is not Null, the option is automatically set to an stocked secondary antibody for the primary antibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankSecondaryAntibodyDilutionFactor
The dilution ratio of BlankSecondaryAntibody. BlankSecondaryAntibody is always diluted in the SecondaryAntibodyDiluent. Either BlankSecondaryAntibodyDilutionFactor or BlankSecondaryAntibodyVolume should be provided but not both.
Default Calculation: If Blank is not Null, for IndirectELISA, IndirectSandwichELISA, IndirectCompetitiveELISA, automatically set to 0.001 (1:1,000).
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or Null.
Programmatic Pattern: (RangeP[0, 1] | Automatic) | Null
BlankSecondaryAntibodyVolume
The volume of BlankSecondaryAntibody added into the corresponding well of the assay plate. BlankSecondaryAntibodyVolume is used as an alternative to BlankSecondaryAntibodyDilutionFactor. During antibody preparation, a master mix will be made for antibody dilution, and the diluted Antibodies will be aliquoted into each well.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: RangeP[0*Microliter, 200*Microliter] | Null
BlankCoatingVolume
The amount of Blank that is aliquoted into the ELISAPlate.
Default Calculation: If Blank is not Null, and Method is DirectELISA or IndirectELISA, CoatingVolume is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankReferenceAntigenCoatingVolume
The amount of diluted BlankReferenceAntigen that is aliquoted into the assay plate, in order for the BlankReferenceAntigen to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectCompetitiveELISA or IndirectCompetitiveELISA, CoatingVolume is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankCoatingAntibodyCoatingVolume
The amount of diluted BlankCoatingAntibody that is aliquoted into the ELISAPlate, in order for the BlankCoatingAntibody to be adsorbed to the surface of the well.
Default Calculation: If Method is FastELISA, the Option is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankCaptureAntibodyCoatingVolume
The amount of diluted BlankCaptureAntibody that is aliquoted into the ELISAPlate, in order for the BlankCaptureAntibody to be adsorbed to the surface of the well.
Default Calculation: If Method is DirectSandwichELISA or IndirectSandwichELISA, the option is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankBlockingVolume
The amount of BlankBlockingBuffer that is aliquoted into the appropriate wells of the ELISAPlate.
Default Calculation: If Blocking is True, the option is automatically set to 200 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 300 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 300*Microliter] | Automatic) | Null
BlankAntibodyComplexImmunosorbentVolume
The volume of the BlankAntibodyComplex to be loaded on the ELISAPlate. In DirectCompetitiveELISA and IndirectCompetitiveELISA, this step enables the free primary antibody to bind to the ReferenceAntigen coated on the plate. In FastELISA, this step enables the PrimaryAntibody-TargetAntigen-CaptureAntibody complex to bind to the CoatingAntibody on the plate.
Default Calculation: If the Method is set to DirectCompetitiveELISA, IndirectCompetitiveELISA, or FastELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankImmunosorbentVolume
The volume of the Blank to be loaded on the ELISAPlate for the target antigen to bind to the capture antibody in DirectSandwichELISA and IndirectSandwichELISA.
Default Calculation: If the Method is set to DirectSandwichELISA and IndirectSandwichELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankPrimaryAntibodyImmunosorbentVolume
The volume of the BlankPrimaryAntibody to be loaded on the ELISAPlate for Immunosorbent assay.
Default Calculation: If the Method is set to DirectELISA, IndirectELISA, DirectSandwichELISA, or IndirectSandwichELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankSecondaryAntibodyImmunosorbentVolume
The volume of the Secondary Antibody to be loaded on the ELISAPlate for immunosorbent step.
Default Calculation: If the Method is set to IndirectELISA, IndirectSandwichELISA, and IndirectCompetitiveELISA, the option is automatically set to 100 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankSubstrateSolution
Defines the substrate solution for the Blank.
Default Calculation: If enzyme is Horseredish Peroxidase, the option will be automatically set to Model[Sample,"ELISA TMB Stabilized Chromogen"]. If enzyme is Alkaline Phosphatase, then the option is automatically set to Model[Sample,"AP Substrate PNPP Solution"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankStopSolution
The reagent that is used to stop the reaction between the enzyme and its substrate.
Default Calculation: If enzyme is Horseradish Peroxidase, then the option is automatically set to Model[Sample,"ELISA HRP-TMB Stop Solution"]. If enzyme is Alkaline Phosphatase, then the option is automatically set to Model[Sample,"AP Substrate Stop Solution"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankSubstrateSolutionVolume
The volume of BlankSubstrateSolution to be added to the corresponding well.
Default Calculation: If SubstrateSolution is populated, then the option is automatically set to 100ul.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
BlankStopSolutionVolume
The volume of BlankStopSolution to be added to the corresponding well.
Default Calculation: If Model[Sample,"ELISA TMB Stabilized Chromogen"] or Model[Sample,"ELISA HRP-TMB Stop Solution"] is selected, the StopSolutionVolume is automatically set to 100ul.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 200 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 200*Microliter] | Automatic) | Null
Assay Plate
ELISAPlate
The assay plate each sample, standard, and blank will be loaded into and the immunosorbent assay will take place. This plate can be pre-coated and blocked in advance.
Default Calculation: The option will be automatically set to an optical, flat-bottom 96 well plate made with polystyrene. If Samples are pre-coated to the a plate, the option will be automatically set to this plate.
Pattern Description: An object of type or subtype Model[Container, Plate] or Object[Container, Plate] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Container, Plate], Object[Container, Plate]}] | _String) | Automatic
SecondaryELISAPlate
The second assay plate, if needed.
Default Calculation: If the total number of Sample (including spiked samples and dilutions), standards, and blanks, times NumberOfReplications exceeds 96, then the option will be automatically set to an optical, flat-bottom 96 well plate made with polystyrene. If Samples are pre-coated to two plates, the option will be automatically set to the second plate.
Pattern Description: An object of type or subtype Model[Container, Plate] or Object[Container, Plate] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Plate], Object[Container, Plate]}] | _String) | Automatic) | Null
ELISAPlateAssignment
Specifies the placement of samples and their corresponding spikes, dilutions, and antibodies in the first ELISAPlate.
Default Calculation: Samples are automatically assigned according to the input sequence (UnknownSamples,Standards,Blanks), to the ELISAPlate from the A1 position, consecutively, in a column-wise fashion. Each sample and their dilution curve will be placed next to each other, then each replicates are placed next to each other. When samples are pre-coated onto the plate, they are arranged from the A1 position, consequtively, in a column-wise fashion,regardless of the input sequence. If the first plate cannot accommodate all the samples, the samples will automatically be assigned to the SecondaryELISAPlate. Due to possible plate-to-plate variations, when two plates are needed, users are recommended to rearrange the samples so that the samples, standards, and blanks that are detected using the same conditions (i.e. same experiment) should be placed in the same plate. If impossible, additional standards and blanks should be included so that each sample has a corresponding set of standards and blanks on the same plate.
Pattern Description: List of one or more {Type, Sample, Spike, SpikeDilutionFactor, SampleDilutionFactors, CoatingAntibody, CoatingAntibodyDilutionFactor, CaptureAntibody, CaptureAntibodyDilutionFactor, ReferenceAntigen, ReferenceAntigenDilutionFactor, PrimaryAntibody, PrimaryAntibodyDilutionFactor, SecondaryAntibody, SecondaryAntibodyDilutionFactor, CoatingVolume, BlockingVolume, SampleAntibodyComplexImmunosorbentVolume, SampleImmunosorbentVolume, PrimaryAntibodyImmunosorbentVolume, SecondaryAntibodyImmunosorbentVolume, SubstrateSolution, StopSolution, SubstrateSolutionVolume, StopSolutionVolume} entries.
Programmatic Pattern: {{Unknown | Spike | Standard | Blank, ObjectP[{Model[Sample], Object[Sample]}] | _String, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, {RangeP[0, 1]..}, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, ObjectP[{Model[Sample], Object[Sample]}] | _String, RangeP[0, 1] | Null, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 300*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, ObjectP[{Model[Sample], Object[Sample]}] | _String, ObjectP[{Model[Sample], Object[Sample]}] | _String, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null}..} | Automatic
SecondaryELISAPlateAssignment
Specifies the placement of samples and their corresponding spikes, dilutions, and antibodies in the second ELISAPlate.
Default Calculation: Samples are automatically assigned according to the input sequence (UnknownSamples,Standards,Blanks), to the ELISAPlate from the A1 position, consecutively, in a column-wise fashion. Each sample and their dilution curve will be placed next to each other, then each replicates are placed next to each other. When samples are pre-coated onto the plate, they are arranged from the A1 position, consequtively, in a column-wise fashion,regardless of the input sequence. If the first plate cannot accommodate all the samples, the samples will automatically be assigned to the SecondaryELISAPlate. Due to possible plate-to-plate variations, when two plates are needed, users are recommended to rearrange the samples so that the samples, standards, and blanks that are detected using the same conditions (i.e. same experiment) should be placed in the same plate. If impossible, additional standards and blanks should be included so that each sample has a corresponding set of standards and blanks on the same plate.
Pattern Description: List of one or more {Type, Sample, Spike, SpikeDilutionFactor, SampleDilutionFactors, CoatingAntibody, CoatingAntibodyDilutionFactor, CaptureAntibody, CaptureAntibodyDilutionFactor, ReferenceAntigen, ReferenceAntigenDilutionFactor, PrimaryAntibody, PrimaryAntibodyDilutionFactor, SecondaryAntibody, SecondaryAntibodyDilutionFactor, CoatingVolume, BlockingVolume, SampleAntibodyComplexImmunosorbentVolume, SampleImmunosorbentVolume, PrimaryAntibodyImmunosorbentVolume, SecondaryAntibodyImmunosorbentVolume, SubstrateSolution, StopSolution, SubstrateSolutionVolume, StopSolutionVolume} entries or Null.
Programmatic Pattern: ({{Unknown | Spike | Standard | Blank, ObjectP[{Model[Sample], Object[Sample]}] | _String, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, {RangeP[0, 1]..}, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, ObjectP[{Model[Sample], Object[Sample]}] | _String, RangeP[0, 1] | Null, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Null, RangeP[0, 1] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 300*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null, ObjectP[{Model[Sample], Object[Sample]}] | _String, ObjectP[{Model[Sample], Object[Sample]}] | _String, RangeP[0*Microliter, 200*Microliter] | Null, RangeP[0*Microliter, 200*Microliter] | Null}..} | Automatic) | Null
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and when the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise set to Model[Container, Vessel, "2mL Tube"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[Model[Container]] | Automatic) | Null
Index Matches to: experiment samples
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
Index Matches to: experiment samples
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples