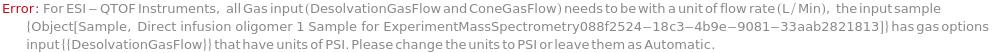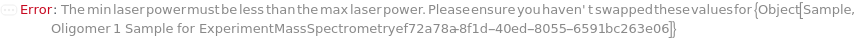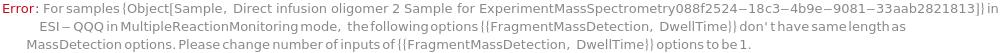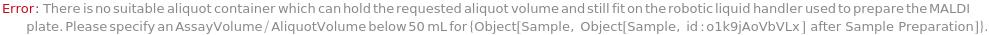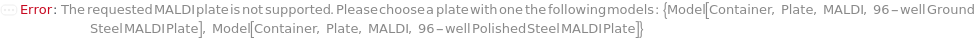General
IonSource
The type of ionization used to create gas phase ions from the molecules in the sample. Electrospray ionization (ESI) produces ions using an electrospray in which a high voltage is applied to a liquid to create an aerosol, and gas phase ions are formed from the fine spray of charged droplets as a result of solvent evaporation and Coulomb fission. In matrix-assisted laser desorption/ionization (MALDI), the sample is embedded in a laser energy absorbing matrix which is then irradiated with a pulsed laser, ablating and desorbing the molecules with minimal fragmentation and creating gas phase ions from the analyte molecules in the sample.
Default Calculation: Is automatically set to ESI or MALDI, respectively, whenever ESI or MALDI specific options or IonSource are specified. If none specified, defaults to MALDI if sample contains DNA oligomers and other synthetic nucleic acid oligomers, and ESI for all other samples.
Pattern Description: ESI or MALDI.
Programmatic Pattern: IonSourceP | Automatic
Instrument
The instrument used to perform the mass spectrometry analysis by ionization of sample and analysis by sequential mass analyzers.
Default Calculation: Is automatically set to the MALDI time-of-flight (TOF) (Model[Instrument, MassSpectrometer, "Microflex LT"]) whenever the IonSource option is set to MALDI and/or MALDI specific options are specified. Otherwise, defaults to ESI-QTOF (Model[Instrument, MassSpectrometer, "Xevo G2-XS QTOF"]).
Pattern Description: An object of type or subtype Model[Instrument, MassSpectrometer] or Object[Instrument, MassSpectrometer]
Programmatic Pattern: ObjectP[{Model[Instrument, MassSpectrometer], Object[Instrument, MassSpectrometer]}] | Automatic
MassAnalyzer
The manner of detection used to resolve and detect molecules. For now, we have 3 mass analyzers available for ExperimentMassSpectrometry. Time-Of-Flight (TOF) analyzer separates the ions by their flight time. QTOF accelerates ions through an elongated flight tube, followed by traveling down the quandrupole analyzer where only ions with selected mass to charge ratio will pass (either a mass range or specific mass value), and then passing a TOF analyzer. QQQ accelerates the ions and selects through two quadrupole analyzers. Both QTOF and QQQ have a collision cell, which optionally fragments the ions, in between the first and the second mass analyzer.
Default Calculation: For MALDI as the ion source, MassAnalyzer is automatically set to TOF detector, as TOF is the only detector that connects to MALDI for now. For ESI ion source, the detector can be TOF/QTOF for the ESI-QTOF study and quadrupole(Q)/triple-quadrupole (QQQ) for ESI-QQQ study. For APCI as the ion source, the analyzer can be quadrupole or triple-quadrupole.
Pattern Description: TOF, QTOF, or TripleQuadrupole.
Programmatic Pattern: (TOF | QTOF | TripleQuadrupole) | Automatic
InjectionType
The type of sample submission method to employ for ESI-QTOF and ESI-QQQ. In DirectInfusion, the sample is directly injected into the instrument, using either a built-in fluidics pump system (ESI-QTOF) or a syringe pump (ESI-QQQ), of the mass spectrometer without the use of any mobile phase. Due to the manual nature of the injection, this is well suited for measuring a small number of samples. Furthermore, this is recommended when high sample volumes of are available (>500 Microliter), the sample does not require cooling, and a quick result is desired. It produces a constant signal over time. FlowInjection works by injecting the sample into a flowing solvent stream using a liquid chromatography autosampler and pumps, in the absence of chromatographic separation. FlowInjection can accomodate up to 2*96 samples and is thus suited for highthrougput analyses. It produces a brief peak of signal at the time that the analyte reaches the detector.
Default Calculation: Is automatically set to Null for MALDI mass spectrometry. If ESI mass spectrometry, is automatically set to FlowInjection if the number of samples is larger than 5, and/or the samples are inside a 96 deep well plate, and/or none of the flow injection options are set, otherwise is set to DirectInfusion.
Pattern Description: DirectInfusion or FlowInjection or Null.
Programmatic Pattern: ((DirectInfusion | FlowInjection) | Automatic) | Null
Analytes
The compounds of interest that are present in the given samples, used to determine the other settings for the Mass Spectrometer (ex. MassDetection).
Default Calculation: If populated, will resolve to the user-specified Analytes field in the Object[Sample]. Otherwise, will resolve to the larger compounds in the sample, in the order of Proteins, Peptides, Oligomers, then other small molecules. Otherwise, set Null.
Pattern Description: List of one or more an object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] entries or Null.
Programmatic Pattern: ({ObjectP[IdentityModelTypes]..} | Automatic) | Null
Index Matches to: experiment samples
IonMode
Indicates if positively or negatively charged ions are analyzed.
Default Calculation: For oligomer samples of the types Peptide, DNA, and other synthetic oligomers, is automatically set to positive ion mode. For other types of samples, defaults to positive ion mode, unless the sample is acid (Acid->True or pKa<=8).
Pattern Description: Negative or Positive.
Programmatic Pattern: IonModeP | Automatic
Index Matches to: experiment samples
MassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded during analysis.
Default Calculation: For MALDI-TOF measurements, is automatically set to ensure there are 3 calibrant peaks which flank the sample's molecular weight in the range. For ESI-QTOF measurements, is automatically set to one of three default mass ranges according to the molecular weight and/or the type of sample (small molecules -> 50-1200 m/z, peptides -> 350 - 2000 m/z, proteins/antibodies -> 500 - 5000 m/z). For ESI-QQQ, the mass range is set by default to be 100 - 1250 m/z if ScanMode is FullScan.
Pattern Description: Multiple or Range or Single.
Programmatic Pattern: (RangeP[5*(Gram/Mole), 2000*(Gram/Mole)] | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 500000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 2000*(Gram/Mole)]..}) | Automatic
Index Matches to: experiment samples
Calibrant
A sample with components of known mass-to-charge ratios (m/z) used to calibrate the mass spectrometer. In the chosen ion polarity mode, the calibrant should contain at least 3 masses spread over the mass range of interest.
Default Calculation: Automatically set based on the sample type and MassDetection. For MALDI, is set to Model[Sample,StockSolution,Standard,"IDT ssDNA Ladder 10-60 nt, 40 ng/uL"] for DNA, and Model[Sample,StockSolution,Standard,"Peptide/Protein Calibrant Mix"] for all others.For ESI, is set to sodium iodide for peptide samples, cesium iodide for intact protein analysis. For other types of samples, is set to cesium iodide if molecular weight is above 2000 Da, to sodium iodide if molecular weight between 1200 and 2000 Da, and to sodium formate for all others (small molecule range). For ESI-QQQ, the default calibrant is polypropylene glycol (PPG) standard sample from SciEX. Where Model[Sample, "id:zGj91a71kXEO"] and Model[Sample, "id:bq9LA0JA1YJz"] are for the positive and negative ion mode, respectively.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
Index Matches to: experiment samples
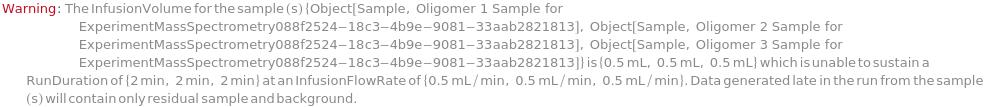
InfusionFlowRate
The flow rate at which the sample is injected into the mass spectrometer through either the DirectInfustion (the fluidics pump system (ESI-QTOF) or the syringe pump system (ESI-QQQ)) or the FlowInjection under a flowing solvent stream using a liquid chromatography autosampler). Note that source settings such as source voltage/temperature and desolvation temperature/flow rate should be adjusted according to the flow rate for improved sensitivity and spray stability.
Default Calculation: In DirectInfusion: Is automatically set to 20 Microliter/Minute and 5 Microliter/Minute for ESI-QTOF and ESI-QQQ, respectively; In FlowInjection; 100 Microliter/Minute for both ESI-QQQ and ESI-QTOF; otherwise is set to Null.
Pattern Description: Greater than or equal to 1 microliter per minute and less than or equal to 600 microliters per minute or Null.
Programmatic Pattern: (RangeP[1*(Microliter/Minute), 600*(Microliter/Minute)] | Automatic) | Null
Index Matches to: experiment samples
ScanTime
The duration of time allowed to pass between each spectral acquisition. Increasing this value improves sensitivity, whereas decreasing this value allows for more data points and spectra to be acquired during the RunDuration.
Default Calculation: Is automatically set to 1 second for ESI-QTOF mass spectrometry. For ESI-QQQ, in MultipleReactionMonitoring, this option is auto resolved by summing up all dwell times of each assay. For FullScan, ScanTime is set to the minimum allowed for the specified MassDetection range (3 microseconds per discrete point in the MassDetection range). For the rest of scan modes, is set to 5 Millisecond (0.005 Second) by default. Otherwise is set to Null.
Pattern Description: Greater than or equal to 0.005 milliseconds and less than or equal to 195 seconds or Null.
Programmatic Pattern: (RangeP[0.005*Millisecond, 195*Second] | Automatic) | Null
Index Matches to: experiment samples
NumberOfReplicates
Number of times each of the input samples should be analyzed using identical experimental parameters.
Pattern Description: Greater than or equal to 2 in increments of 1 or Null.
Programmatic Pattern: GreaterEqualP[2, 1] | Null
MALDI Ionization
NumberOfShots
The total number of times the laser is fired by the MALDI mass spectrometer during data acquisition.
Default Calculation: Is automatically set to 500 for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 50 and less than or equal to 10000 or Null.
Programmatic Pattern: (RangeP[50, 10000] | Automatic) | Null
ShotsPerRaster
The number of repeated laser shots made between each raster movement within a well during a MALDI measurement.
Default Calculation: Is automatically set to 12 for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 10000 or Null.
Programmatic Pattern: (RangeP[1, 10000] | Automatic) | Null
LaserPowerRange
The min and max laser power used during analysis (given as a relative percentage of the mass spectrometer's actual laser power). Adjust this value to increase resolution and reduce noise.
Default Calculation: Automatically set according to sample type and molecular weight of the analyte of interest. For DNA below 3500 Da, this is set to 55-75% and for DNA above 3500 Da to 65-85%. For all other samples it is set to 25-75% if below 6000 Da and 25-90% otherwise.
Pattern Description: A span from anything greater than or equal to 0 percent and less than or equal to 100 percent in increments of 1 percent to anything greater than or equal to 0 percent and less than or equal to 100 percent in increments of 1 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent, 1*Percent] ;; RangeP[0*Percent, 100*Percent, 1*Percent] | Automatic) | Null
Index Matches to: experiment samples
DelayTime
The duration of time that is allowed to pass between the laser pulse and the application of the acceleration voltage in order to control for differences in kinetic energy introduced during the ionization process. Use a longer delay time for samples with a higher molecular weight.
Default Calculation: A delay of 250 ns is used for DNA oligomers. A 150 ns delay is used for all other samples.
Pattern Description: Greater than or equal to 0 nanoseconds and less than or equal to 5000 nanoseconds or Null.
Programmatic Pattern: (RangeP[0*Nanosecond, 5000*Nanosecond] | Automatic) | Null
Index Matches to: experiment samples
CalibrantLaserPowerRange
The min and max laser power used during calibration (given as a relative percentage of the mass spectrometer's actual laser power).
Default Calculation: Is automatically set to use the same laser power range determined for the analyte.
Pattern Description: A span from anything greater than or equal to 0 percent and less than or equal to 100 percent in increments of 1 percent to anything greater than or equal to 0 percent and less than or equal to 100 percent in increments of 1 percent or Null.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent, 1*Percent] ;; RangeP[0*Percent, 100*Percent, 1*Percent] | Automatic) | Null
Index Matches to: experiment samples
CalibrantNumberOfShots
The number of times the mass spectrometer fires the laser during calibration.
Default Calculation: Is automatically set to 100 for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 50 and less than or equal to 10000 or Null.
Programmatic Pattern: (RangeP[50, 10000] | Automatic) | Null
MALDI Sample Preparation
SpottingPattern
Indicates if MALDI plate wells are skipped during spotting to decrease any cross contamination risks; All indicates every well on the plate will be spotted and Spaced indicates every other well is filled.
Default Calculation: Is automatically set to All for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: All or Spaced or Null.
Programmatic Pattern: (SpottingPatternP | Automatic) | Null
SpottingDryTime
The minimum amount of time the samples are left to dry after they have been aliquoted onto the MALDI plate.
Default Calculation: Is automatically set to 15 minutes for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 5 minutes and less than or equal to 1 hour or Null.
Programmatic Pattern: (RangeP[5*Minute, 1*Hour] | Automatic) | Null
MALDIPlate
The plate spotted with samples and calibrants mixed with laser energy absorbing matrix material. The MALDI plate is subsequenctly placed into the mass spectrometer's reduced pressure sample chamber and irradiated with laser pulses to produce gas phase ions that enter the mass spectrometer for mass analysis.
Default Calculation: Is automatically set to Model[Container, Plate, MALDI, "96-well Ground Steel MALDI Plate"] for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: An object of type or subtype Model[Container, Plate], Object[Container, Plate], Model[Container, Plate, MALDI], or Object[Container, Plate, MALDI] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Plate], Object[Container, Plate], Model[Container, Plate, MALDI], Object[Container, Plate, MALDI]}] | _String) | Automatic) | Null
Matrix
The laser-absorbing reagent co-spotted with the input samples in order to assist ionization of the sample.
Default Calculation: Automatically set according to sample type and ion mode. For DNA oligomers and positive ion mode, HPA MALDI matrix is used. Otherwise, the PreferredMALDIMatrix of the calibrant used for this sample is used, and if that is not informed, it defaults to THAP MALDI matrix.
Pattern Description: An object of type or subtype Model[Sample, Matrix] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample, Matrix], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CalibrantMatrix
The laser-absorbing reagent co-spotted with the input calibrants in order to assist ionization of the sample.
Default Calculation: Automatically set to according to the PreferredMatrix field of the calibrant models, else will be resolved to be the same as Matrix.
Pattern Description: An object of type or subtype Model[Sample, Matrix] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample, Matrix], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
SpottingMethod
Indicates if or how to layer the input samples and matrix onto the MALDI plate in order to form sample and calibration spots for analysis.
Default Calculation: Is automatically set to the calibrant's preferred spotting method.
Pattern Description: Sandwich or OpenFace or Null.
Programmatic Pattern: (SpottingMethodP | Automatic) | Null
Index Matches to: experiment samples
SampleVolume
The volume taken from each input sample and aliquoted onto the MALDI plate.
Default Calculation: Is automatically set to 0.8 Microliter for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 2 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 2*Microliter] | Automatic) | Null
Index Matches to: experiment samples
CalibrantVolume
The volume taken from each calibrant sample and aliquoted onto the MALDI plate.
Default Calculation: Is automatically set to 0.8 Microliter for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 0.5 microliters and less than or equal to 2 microliters or Null.
Programmatic Pattern: (RangeP[0.5*Microliter, 2*Microliter] | Automatic) | Null
Index Matches to: experiment samples
MatrixControlScans
Indicates if matrix control samples will be spotted to MALDI plate and investigated as the blank control during the experiment for each different combination of MALDI experiment parameters.
Default Calculation: Is automatically set to True for MALDI mass spectrometry, otherwise is set to Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
ESI Flow Injection
SampleTemperature
The temperature at which the samples are kept in the instrument's autosampler prior to injection. This option can only be changed for ESI-QQQ and ESI-QTOF using FlowInjection as the InjectionType.
Default Calculation: Is automatically set to Ambient for FlowInjection ESI mass spectrometry, otherwise is set to Null.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[5*Celsius, 40*Celsius] | Ambient) | Automatic) | Null
Buffer
The solvent pumped through the flow path, carrying the injected sample to the ionization source where the analytes are ionized via electrospray ionization.
Default Calculation: Is automatically set to Model[Sample,StockSolution,"0.1% FA with 5% Acetonitrile in Water, LCMS-grade"] for FlowInjection ESI mass spectrometry, otherwise is set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
NeedleWashSolution
The solvent used to wash the injection needle before each sample measurement.
Default Calculation: Is automatically set to Model[Sample,StockSolution,"20% Methanol in MilliQ Water"] for FlowInjection ESI mass spectrometry, otherwise is set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or Null.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | Automatic) | Null
InjectionVolume
The physical quantity of sample loaded into the flow path for measurement.
Default Calculation: Is automatically set to 10 Microliter for FlowInjection ESI mass spectrometry (For Both QQQ and QTOF), otherwise is set to Null.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[1*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: experiment samples
ESI-QQQ Direct Infusion
InfusionVolume
The physical quantity of sample loaded into the syringe (for now ESI-QQQ only), when using syringe pump to infusion load the sample.
Default Calculation: Is automatically set to 500 Microliter for DirectInfusion ESI mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 0.05 milliliters and less than or equal to 10 milliliters or Null.
Programmatic Pattern: (RangeP[0.05*Milliliter, 10*Milliliter] | Automatic) | Null
Index Matches to: experiment samples
InfusionSyringe
The syringe used for syringe pump infusion injection (For ESI-QQQ only).
Default Calculation: For ESI-QQQ, is automatically resolved based on the infusion volume: (0.01 mL ~ 0.99 mL) -> Model[Container, Syringe, "1mL All-Plastic Disposable Syringe"]; (1.01 mL ~ 2.99 mL) ->Model[Container, Syringe, "3mL Sterile Disposable Syringe"];(3.00 mL,4.99 mL)->Model[Container, Syringe, "5mL Sterile Disposable Syringe"];(5.0mL,9.9mL)->Model[Container, Syringe, "10mL Syringe"]. Otherwise is set to Null.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
Method Information
RunDuration
The duration of time for which spectra are acquired for the sample currently being injected.
Default Calculation: Is automatically set to 1 minute for ESI-QTOF and ESI-QQQ mass spectrometry, otherwise is set to Null.
Pattern Description: Greater than or equal to 0.1 minutes and less than or equal to 60 minutes or Null.
Programmatic Pattern: (RangeP[0.1*Minute, 60*Minute] | Automatic) | Null
Index Matches to: experiment samples
ESI Ionization
ESICapillaryVoltage
This option (also known as Ion Spray Voltage) indicates the absolute voltage applied to the tip of the stainless steel capillary tubing in order to produce charged droplets. Adjust this voltage to maximize sensitivity. For ESI-QTOF, most compounds are optimized between 0.5 and 3.2 kV in ESI positive ion mode and 0.5 and 2.6 kV in ESI negative ion mode. For ESI-QQQ this parameter is 5.5 kV for positive ion mode and -4.5 kV for negative ion mode, for standard flow UPLC a value of 0.5 kV is typically best for maximum sensitivity. Thie parameter can be altered according to sample type. For low flow applications, best sensitivity will be achieved with a relatively high value in ESI positive (e.g. 3.0 kV for ESI-QTOF and 5.5 or -4.5 kV for ESI-QQQ positive and negative, respectively).
Default Calculation: For ESI-QTOF: Is automatically set according to the flow rate: For ESI-QTOF: 0-0.02 ml/min -> 3.0 kV (Positive) or 2.8 kV (Negative), 0.021-0.1 ml/min -> 2.5 kV, 0.101 - 0.3 ml/min -> 2.0 kV, 0.301 - 0.5 ml/min -> 1.5 kV, > 0.5 ml/min -> 1.0 kv . For ESI-QQQ: < 0.02 ml/min -> 5.5 kV or -4.5 kV, 0.02-0.1 ml/min -> 4.5 kV or -4.0 KV, >0.1 ml/min -> 4.0 kV and -4.0 kV, for positive and negative IonMode respectively.
Pattern Description: Greater than or equal to -4.5 kilovolts and less than or equal to 5.5 kilovolts or Null.
Programmatic Pattern: (RangeP[-4.5*Kilovolt, 5.5*Kilovolt] | Automatic) | Null
Index Matches to: experiment samples
DeclusteringVoltage
For ESI-QTOF, indicates the voltage between the ion block (the reduced-pressure chamber of the source block) and the stepwave ion guide (the optics before the quadrupole mass analyzer). This voltage attracts charged ions from the capillary tip into the ion block leading into the mass spectrometer. For ESI-QTOF, this voltage is typically set to 25-100 V and its tuning has little effect on sensitivity. For ESI-QQQ, this controls the voltage applied between the orifice (where ions enter the mass spectrometer) and the ion guide to prevent the ionized small particles from aggregating together. It's normally optimized between -300 to 300 V, and its sensitivity depends on chemical composition and charge state.
Default Calculation: Is automatically set to any specified method; otherwise, for ESI-QTOF, is set to 40 Volt; and for ESI-QQQ, is set to 90 Volt and -90 respectively for positive and negative ion mode.
Pattern Description: Greater than or equal to -400 volts and less than or equal to 400 volts or Null.
Programmatic Pattern: (RangeP[-400*Volt, 400*Volt] | Automatic) | Null
Index Matches to: experiment samples
StepwaveVoltage
This is a unique option for ESI-QTOF. It indicates the voltage offset between the 1st and 2nd stage of the ion guide which leads ions coming from the sample cone towards the quadrupole mass analyzer. This voltage normally optimizes between 25 and 150 V and should be adjusted for sensitivity depending on compound and charge state. For multiply charged species it is typically set to to 40-50 V, and higher for singly charged species. In general higher cone voltages (120-150 V) are needed for larger mass ions such as intact proteins and monoclonal antibodies. It also has greatest effect on in-source fragmentation and should be decreased if in-source fragmentation is observed but not desired.
Default Calculation: Is automatically set according to the sample type (proteins, antibodies and analytes with MW > 2000 -> 120 V, DNA and synthetic nucleic acid oligomers -> 100 V, all others (including peptides and small molecules) -> 40 V).
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 200 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 200*Volt] | Automatic) | Null
Index Matches to: experiment samples
SourceTemperature
The temperature setting of the source block. Heating the source block discourages condensation and decreases solvent clustering in the reduced vacuum region of the source. For ESI-QTOF, This temperature setting is flow rate and sample dependent. Typical values are between 60 to 120 Celsius. For thermally labile analytes, a lower temperature setting is recommended. For ESI-QQQ, this values is set to 150 Celsius cannot be changed.
Default Calculation: For ESI-QTOF: is automatically set according to the flow rate (0-0.02 ml/min -> 100 Celsius, 0.021-0.3 ml/min -> 120 Celsius, >0.3 ml/min -> 150 Celsius). For ESI-QQQ, this value is locked to be 150 Celsius.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: (RangeP[$AmbientTemperature, 150*Celsius] | Automatic) | Null
Index Matches to: experiment samples
DesolvationTemperature
The temperature setting for the ESI desolvation heater that controls the nitrogen gas temperature used for solvent evaporation to produce single gas phase ions from the ion spray. Similarly to DesolvationGasFlow, this setting is dependant on solvent flow rate and composition. A typical range is from 150 to 650 Celsius for ESI-QTOF and 350 to 600 Celsius for ESI-QQQ.
Default Calculation: Is automatically set according to the flow rate: For ESI-QTOF (0-0.02 ml/min -> 200 Celsius, 0.021-0.1 ml/min -> 350 Celsius, 0.101-0.3 -> 450 Celsius, 0.301->0.5 ml/min -> 500 Celsius, >0.500 ml/min -> 600 Celsius); For ESI-QQQ (0-0.02 ml/min -> 350 Celsius, 0.021-0.1 ml/min -> 400 Celsius, 0.101-0.3 -> 450 Celsius, 0.301->0.5 ml/min -> 500 Celsius, >0.500 ml/min -> 600 Celsius); .
Pattern Description: Greater than or equal to 0 degrees Celsius and less than or equal to 750 degrees Celsius or Null.
Programmatic Pattern: (RangeP[0*Celsius, 750*Celsius] | Automatic) | Null
Index Matches to: experiment samples
DesolvationGasFlow
The nitrogen gas flowing around the electrospray inlet capillary in order to desolvate and nebulize analytes. Similarly to DesolvationTemperature, the ideal setting is dependent on solvent flow rate and composition. When MassAnalyzer is QQQ, this value is in terms of pressure (PSI). When MassAnalyzer is QTOF, this value is in terms of flow rate (Liter/Hour).
Default Calculation: Is automatically set according to the flow rate: for ESI-QTOF (0-0.02 ml/min -> 600 L/H, 0.021-0.3 ml/min -> 800 L/H, 0.301-0.5 -> 1000 L/H, >0.500 ml/min -> 1200 L/H); and (0-0.02 ml/min -> 20 PSI, 0.02-0.3 ml/min -> 40 PSI, 0.301-0.500 ml/min -> 60 PSI, >0.500 ml/min -> 80) for ESI-QQQ.
Pattern Description: Greater than or equal to 55 liters per hour and less than or equal to 1200 liters per hour or greater than or equal to 0 pounds
‐force per inch squared and less than or equal to 85 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[55*(Liter/Hour), 1200*(Liter/Hour)] | RangeP[0*PSI, 85*PSI]) | Automatic) | Null
Index Matches to: experiment samples
ConeGasFlow
The nitrogen gas flow ejected around the sample inlet cone (the spherical metal plate acting as a first gate between the sprayer and the reduced pressure chamber, the ion block). This gas flow is used to minimize the formation of solvent ion clusters. It also helps reduce adduct ions and directing the spray into the ion block while keeping the sample cone clean. The same parameter is referred to as Curtain Gas Pressure for ESI-QQQ. Typical values are between 0 and 150 L/h for ESI-QTOF or 20 to 55 PSI for ESI-QQQ.
Default Calculation: Is automatically set to 50 Liter/Hour for ESI-QTOF and 50 PSI for ESI-QQQ, and is set to Null in MALDI-TOF. Is not recommended to set to a smaller value of 40 PSI in ESI-QQQ, due to potential deposition of the sample inside the instrument that will lead to contamination.
Pattern Description: Greater than or equal to 0 liters per hour and less than or equal to 300 liters per hour or greater than or equal to 20 pounds
‐force per inch squared and less than or equal to 55 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[0*(Liter/Hour), 300*(Liter/Hour)] | RangeP[20*PSI, 55*PSI]) | Automatic) | Null
Index Matches to: experiment samples
IonGuideVoltage
This option (also known as Entrance Potential (EP)) is a unique option of ESI-QQQ. This parameter indicates electric potential applied to the Ion Guide in ESI-QQQ, which guides and focuses the ions through the high-pressure ion guide region.
Default Calculation: Is automatically set to 10 V for positive ions, or
–10 V for negative ions in ESI-QQQ, and can be changed between 2-15 V in both positive and negative mode. This value is set to Null in ESI-QTOF.
Pattern Description: Greater than or equal to -15 volts and less than or equal to -2 volts or greater than or equal to 2 volts and less than or equal to 15 volts or Null.
Programmatic Pattern: ((RangeP[-15*Volt, -2*Volt] | RangeP[2*Volt, 15*Volt]) | Automatic) | Null
Index Matches to: experiment samples
MALDI Mass Analysis
AccelerationVoltage
The voltage applied to the MALDI plate in order to accelerate ions towards the detector. Increase this voltage to increase the sensitivity or decrease it to increase the resolution.
Default Calculation: Automatically set to 19.5 kV for MALDI mass spectrometry, otherwise set to Null.
Pattern Description: Greater than or equal to 0.1 kilovolts and less than or equal to 20 kilovolts or Null.
Programmatic Pattern: (RangeP[0.1*Kilovolt, 20*Kilovolt] | Automatic) | Null
Index Matches to: experiment samples
GridVoltage
The voltage applied to a secondary plate above the MALDI plate in order to create a gradient such that ions with lower initial kinetic energies are accelerated faster than samples with higher kinetic energies which have drifted farther from the MALDI plate. Use a lower grid voltage for samples with a higher molecular weight.
Default Calculation: Automatically set to 18.15 kV for MALDI mass spectrometry, otherwise set to Null.
Pattern Description: Greater than or equal to 0.1 kilovolts and less than or equal to 20 kilovolts or Null.
Programmatic Pattern: (RangeP[0.1*Kilovolt, 20*Kilovolt] | Automatic) | Null
Index Matches to: experiment samples
LensVoltage
The voltage applied to the electrostatic ion focusing lens located at the entrance of the mass analyser.
Default Calculation: Automatically set to 7.8 kV for MALDI mass spectrometry, otherwise set to Null.
Pattern Description: Greater than or equal to 0.05 kilovolts and less than or equal to 10 kilovolts or Null.
Programmatic Pattern: (RangeP[0.05*Kilovolt, 10*Kilovolt] | Automatic) | Null
Index Matches to: experiment samples
Gain
The signal amplification factor applied to the detector in MALDI-TOF analysis. A gain factor of one corresponds to the lowest voltage applied to the electron multiplier. Larger values can increase signal intensity, but may cause saturation and decreased signal to noise.
Default Calculation: Is automatically set to two
Pattern Description: Greater than or equal to 1. and less than or equal to 10 or Null.
Programmatic Pattern: (RangeP[1., 10.] | Automatic) | Null
Index Matches to: experiment samples
Tandem Mass Spectrometry
ScanMode
The acquisition and selection sequence when MassAnalyzer is TripleQuadrupole. Different scan modes will apply selections to the first (MS1) and third (MS2) quadrupole at specific times. In Full Scan Mode, the entire MS1 range is scanned and fragmentation is off. In SelectedIonMonitoring, select MS1 masses (per the MassDetection option) are monitored and measured without fragmentation. In PrecursorIonScan mode, fragmentation is on, and fragment ion is selected (per the FragmentMassDetection option), while MS1 masses are scanned across a range (MassDetection). In NeutralIonLoss mode, both MS1 and MS2 masses are scanned, in order to track specific MS1/MS2 combinations for neutral ion loss. In ProductIonScan, an MS1 mass is selected (MassDetection) and a range of MS2 mass is scanned in order to survey fragmentation patterns of the parent mass. In MultipleReactionMonitoring mode, both MS1 and MS2 are selected with specific intact and fragment ion pairs are monitored. ScanMode is automatically set to FullScan (no fragmentation of the sample, collecting full mass range scan).
Default Calculation: For ESI-QQQ this value will be automatically resolved based-on tandem mass related input. For ESI-QTOF and MALDI-TOF, this will be resolved to Null.
Pattern Description: FullScan, SelectedIonMonitoring, PrecursorIonScan, NeutralIonLoss, ProductIonScan, or MultipleReactionMonitoring or Null.
Programmatic Pattern: (MassSpecScanModeP | Automatic) | Null
Index Matches to: experiment samples
FragmentMassDetection
The mass scan range for the second mass analyzer (MS2) in the Tandem MS study. The second mass anaylzer screens and scans the ion after the incoming ions have been fragmented by collision cells. This option can be set at one specific mass value (mass selection mode), or at a mass range (mass scan mode). This option will be checked to match the corresponding ScanMode.
Default Calculation: Is automatically resolved as Null for FullScan and SelectedIonMonitoring mode. Is set to 5 - 1250 m/z by default for ProductionIonScan modes and resolved to be the same as MassDetection in NeutralIonLoss model, and resolved based on MassDetection or 500 m/z for PrecursorIonScan and MultipleReactionMonitoring mode.
Pattern Description: Multiple or Range or Single or Null.
Programmatic Pattern: ((RangeP[5*(Gram/Mole), 2000*(Gram/Mole)] | RangeP[5*(Gram/Mole), 2000*(Gram/Mole)] ;; RangeP[5*(Gram/Mole), 2000*(Gram/Mole)] | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..}) | Automatic) | Null
Index Matches to: experiment samples
MassTolerance
This options indicates the step size of both MS1 and MS2 when both or either one of them are set in mass selection mode. This value indicates the mass range used to find a peak with twice the entered range. For example, for a mass range 100 Da to 200 Da and step size 0.1, the instrument scans 99.95 to 100.05 (records as value 100), 100.05 to 101.15 (records as value 101) and 199.95 to 200.05 (records as value 200).
Default Calculation: This option will be set to Null if using MALDI-TOF and ESI-QTOF. For ESI-QQQ, if both of the mass anaylzer are in mass selection mode (SelectedIonMonitoring and MultipleReactionMonitoring mode), this option will be auto resolved to Null. In all other mass scan modes in ESI-QQQ, this option will be automatically resolved to 0.1 g/mol.
Pattern Description: Greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or Null.
Programmatic Pattern: (RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | Automatic) | Null
Index Matches to: experiment samples
Fragment
Determines whether to have ions dissociate upon collision with neutral gas species and to measure the resulting product ions. Also known as tandem mass spectrometry or MS/MS (as opposed to MS).
Default Calculation: For ESI-QQQ, this option will be automatically resolved by the corresponding scan modes (False for FullScan and SelectedIonMonitoring Method, True for PrecursorIonScan, NeutralIonLoss, ProductIonScan and MultipleReactionMonitoring mode). For ESI-QTOF and MALDI-TOF this options will be resolved to False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
CollisionEnergy
The potential used in the collision cell to fragment the incoming ions. Changing the collision energy will change the fragmentation pattern of the incoming ion. High collision energy gives higher ion intensities but the mass patterns will also be more complex. Low collision energy gives simpler mass patterns with lower intensity.
Default Calculation: Is automatically set to 30 V (Positive) and -30 V (Negative) if the collision option is True. This value is Null when Collision option is False and when scan mode is set to MultipleReactionMonitoring.
Pattern Description: Greater than or equal to 5 volts and less than or equal to 180 volts or greater than or equal to -180 volts and less than or equal to 5 volts or list of one or more greater than or equal to 5 volts and less than or equal to 180 volts or greater than or equal to -180 volts and less than or equal to 5 volts entries or Null.
Programmatic Pattern: (((RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | {(RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt])..}) | Automatic) | Null
Index Matches to: experiment samples
CollisionCellExitVoltage
Also known as the Collision Cell Exit Potential (CXP). This value focuses and accelerates the ions out of collision cell (Q2) and into 2nd mass analyzer (MS 2). This potential is tuned to ensure successful ion acceleration out of collision cell and into MS2, and can be adjusted to reach the maximal signal intensity. This option is unique to ESI-QQQ for now, and only required when Fragment ->True and/or in ScanMode that achieves tandem mass feature (PrecursorIonScan, NeutralIonLoss,ProductIonScan,MultipleReactionMonitoring). For non-tandem mass ScanMode (FullScan and SelectedIonMonitoring) and other massspectrometer (ESI-QTOF and MALDI-TOF), this option is resolved to Null.
Default Calculation: Is automatically set to 15 V (Positive mode) or -15 V (Negative mode) if the collision option is True and using QQQ as the mass analyzer.
Pattern Description: Greater than or equal to -55 volts and less than or equal to 55 volts or Null.
Programmatic Pattern: (RangeP[-55*Volt, 55*Volt] | Automatic) | Null
Index Matches to: experiment samples
DwellTime
The duration of time for which spectra are acquired at the specific mass detection value for SelectedIonMonitoring and MultipleReactionMonitoring mode in ESI-QQQ.
Default Calculation: Is automatically set to 200 microsecond for ESI-QQQ mass spectrometry at SelectedIonMonitoring mode, otherwise is set to Null.
Pattern Description: Greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds entries or Null.
Programmatic Pattern: ((RangeP[5*Millisecond, 2000*Millisecond] | {RangeP[5*Millisecond, 2000*Millisecond]..}) | Automatic) | Null
Index Matches to: experiment samples
NeutralLoss
A neutral loss scan is performed on ESI-QQQ instrument by scanning the sample through the first quadrupole (Q1). The ions are then fragmented in the collision cell. The second mass analyzer is then scanned with a fixed offset to MS1. This option represents the value of this offset.
Default Calculation: Is set to 500 g/mol if using scan mode, and is Null in other modes.
Pattern Description: Greater than 0 grams per mole or Null.
Programmatic Pattern: (GreaterP[0*(Gram/Mole)] | Automatic) | Null
Index Matches to: experiment samples
MultipleReactionMonitoringAssays
In ESI-QQQ, the ion corresponding to the compound of interest is targetted with subsequent fragmentation of that target ion to produce a range of daughter ions. One (or more) of these fragment daughter ions can be selected for quantitation purposes. Only compounds that meet both these criteria, i.e. specific parent ion and specific daughter ions corresponding to the mass of the molecule of interest are detected within the mass spectrometer. The mass assays (MS1/MS2 mass value combinations) for each scan, along with the CollisionEnergy and DwellTime (length of time of each scan).
Default Calculation: Need to fill in order to use Multiple Reaction Monitoring mode. Will auto switch to Selected Reaction Monitoring mode if only one mass assay/dwell time is input. Default dwell time is 200 micro second.
Pattern Description: List of one or more {Mass Selection Values, CollisionEnergies, Fragment Mass Selection Values, Dwell Times} entries or Null.
Programmatic Pattern: ({{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..} | Automatic) | Null
Index Matches to: experiment samples
Post Experiment
CalibrantStorageCondition
For each calibrants used in the experiment, the non-default condition under which the calibrants should be stored after the protocol is complete. If not set, the storage condition will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples
Model Input
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 300 Microliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((Null | (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All)) | Automatic) | Null
Index Matches to: experiment samples
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise, automatically set to Model[Container, Vessel, "HPLC vial (high recovery)"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: ((Null | ObjectP[Model[Container]]) | Automatic) | Null
Index Matches to: experiment samples