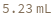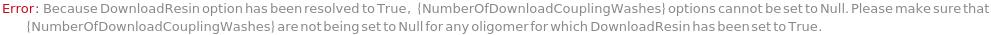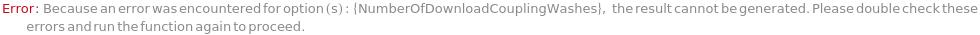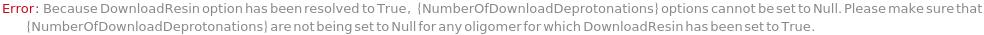ExperimentPeptideSynthesis
ExperimentPeptideSynthesis[OligomerModels]⟹Protocol
uses a oligomer OligomerModels to create a Peptide Synthesis Protocol and associated synthesis cycles methods.
Peptide synthesis is the production of peptides, where multiple amino acids are linked via amide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl group of one amino acid to the amino group of another. This can be achieved with classical solution phase synthesis or solid phase synthesis, which is used in this experiment. In solid phase synthesis, amino acids are covalently bound to a solid support material and synthesised step-by-step in a single reaction vessel utilising selective protecting group chemistry. Peptides are attached to the solid support from their C terminus. Peptide synthesis occurs by coupling the carboxyl group of the incoming amino acid to the N-terminus of the growing peptide chain. This C-to-N synthesis is opposite from protein biosynthesis, during which the N-terminus of the incoming amino acid is linked to the C-terminus of the protein chain (N-to-C). The benefit of the solid support is the isolation of synthesis sites, making interference between growing chains impossible. The use of protecting groups is essential to avoid undesirable side reactions during this process. The most common protecting groups for amino groups used in peptide synthesis are 9-fluorenylmethyloxycarbonyl group (Fmoc), which is used in this experiment, and t-butyloxycarbonyl (Boc). Peptide synthesis allows the production of specific sequences facilitating the study protein function and the development of antibiotic drugs, antibodies and peptide biomaterials. ExperimentTotalProteinQuantification, ExperimentMassSpectrometry and ExperimentHPLC with fraction analysis can be run on the results of this experiment to measure the yield, verify the molecular weight and measure percent purity of the product.
Experimental Principles

Figure 1.1: Procedural overview of a PeptideSynthesis experiment. Step 1: Define the amino acid sequence to be synthesized, including any modifications, by providing a Model[Sample], a Model[Molecule,Oligomer], a Sequence (Peptide["LysTyrValAlaMet"]), Strand (ToStrand[Peptide["LysTyrValAlaMetTyr"]]) or Structure (ToStructure["LysTyrValAlaMetTyrAla", Polymer -> Peptide]). Step 2: The first monomer is loaded to the resin Step 3: The monomers are then expanded with a cycle of a) removal of the Fmoc protecting group from the resin linker or the last amino acid that is currently on the resin, b) the activation of the monomer and c) the addition on the monomer to the resin or resin with linked strand. Step 4: The peptide is liberated from the resin and side chain protecting groups are removed. Step 5: The peptide is purified by using a solution where the desired peptide strands are insoluble and impurities are soluble. This results in the peptides crashing out of solution. Step 6: The peptide is then resuspended in a resuspension buffer.
Instrumentation
Symphony X

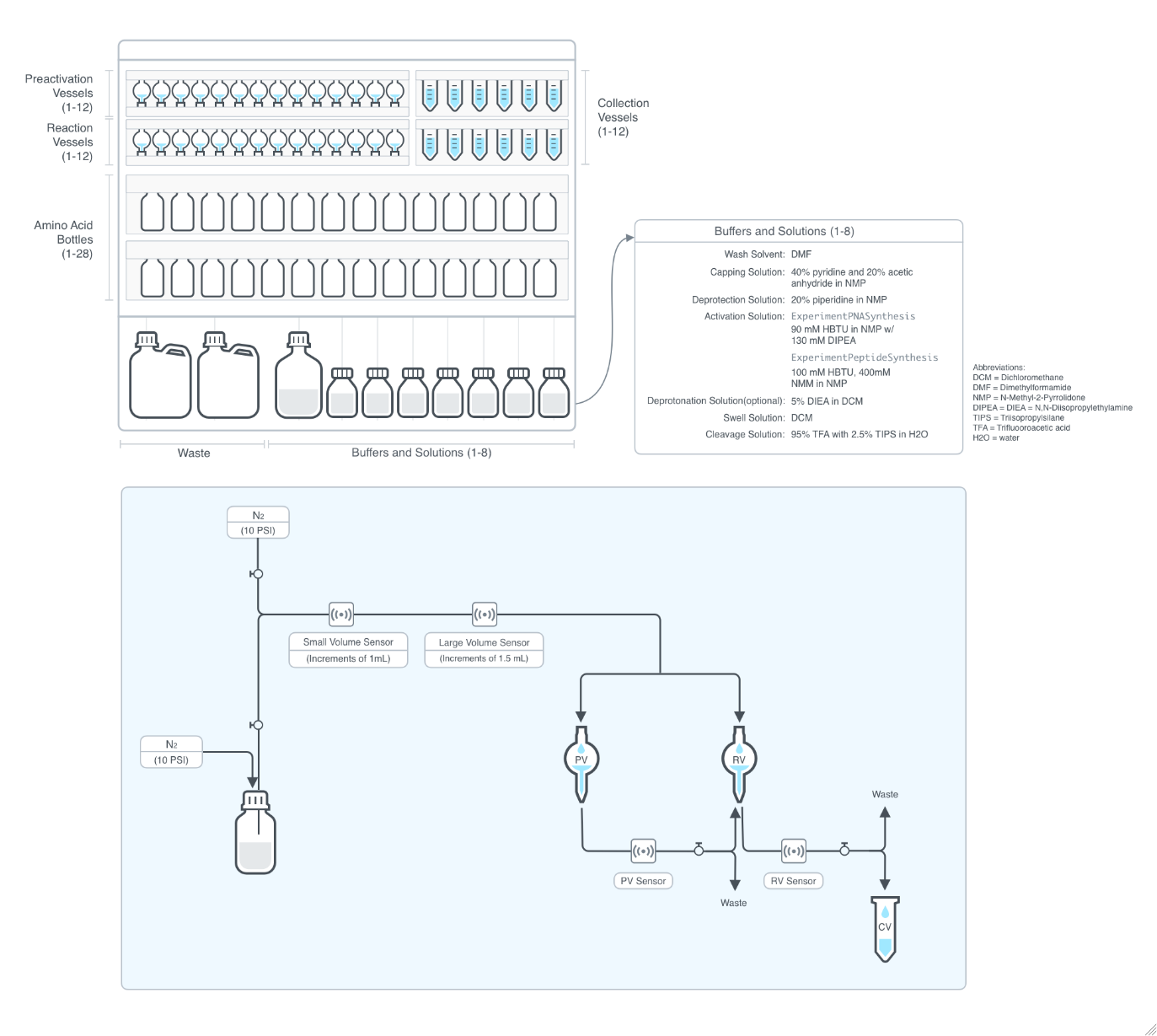
Figure 2.1.1: A system of pre-activation vessels (PV), reaction vessels (RV), amino acid bottles, buffers and solutions, waste and collection vessels (CV) connected by channels. The flow in the channels is controlled by Nitrogen gas and the volume of the liquid deliveries are verified with optical sensors.
Experiment Options
Protocol
Instrument
Pattern Description: An object of type or subtype Model[Instrument, PeptideSynthesizer] or Object[Instrument, PeptideSynthesizer]
Programmatic Pattern: ObjectP[{Model[Instrument, PeptideSynthesizer], Object[Instrument, PeptideSynthesizer]}]
SynthesisStrategy
Scale
Pattern Description: Greater than or equal to 5 micromoles and less than or equal to 100 micromoles in increments of 2.5 micromoles.
TargetLoading
Pattern Description: Greater than or equal to 60 micromoles per gram and less than or equal to 200 micromoles per gram.
Resin
Default Calculation: Will resolve automatically to the undownloaded Resin Model[Sample,"Rink Amide ChemMatrix"] if DownloadResin is set to True. If DownloadResin is set to False, the user is required to specify an appropriate downloaded resin.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
DownloadResin
Indicates if an undownloaded resin will be used and resin download will be performed as the first cycle of the synthesis.
Swelling
SwellSolution
The model or sample object to be used to swell the resin prior to the start of the synthesis or resin download.
Default Calculation: Automatic will resolve to Model[Sample, "Dichloromethane, Reagent Grade"] if SwellResin has been set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
SwellResin
SwellTime
SwellVolume
Pattern Description: Greater than or equal to 2.5 milliliters and less than or equal to 25 milliliters or Null.
NumberOfSwellCycles
Washing
WashSolution
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
WashVolume
Default Calculation: Will automatically resolve to 4 Milliliter plus an additonal .2 Milliliter for each 1 Micromole of synthesis Scale, rounded up to the nearest .5 Milliliter.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters.
Deprotection
DeprotectionSolution
The model or sample object that represents the deprotection solution that will be used to remove protecting groups from the growing strand during the deprotection step of a synthesis or download resin cycle.
Default Calculation: Will resolve to Model[Sample, StockSolution, "Deprotection Solution (20% piperidine in NMP)"] for Fmoc synthesis and Model[Sample, StockSolution, "95% TFA 5% m-cresol"] for Boc synthesis.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
InitialDeprotection
Default Calculation: The initial deprotection will only occur if DownloadResin is set to True as all other cycles already include a deprotection step prior to their coupling steps.
FinalDeprotection
Indicates if a final deprotection step is will be done as part of the last synthesis cycle prior to the start of cleavage or resin storage.
DeprotectionVolume
The volume of DeprotectionSolution added to each reaction vessel during the deprotection step of a synthesis cycle.
Default Calculation: Automatic will resolve to 3 Milliliter plus an additonal .2 Milliliter for each Micromole of synthesis Scale, rounded up to the nearest .5 Milliliter
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters.
DeprotectionTime
The amount of time that each reaction vessel is exposed to the deprotection solution during each deprotection step of a synthesis cycle.
NumberOfDeprotections
The number of times that each reaction vessel will be deprotected per deprotection step of a synthesis cycle.
NumberOfDeprotectionWashes
The number of WashSolution washes each reaction vessel will undergo after the deprotection step of the synthesis cycle.
Deprotonation
DeprotonationSolution
The model or sample object that represents the DeprotonationSolution that will be used to neutralize the resin prior to the coupling step.
Default Calculation: Will automatically resolve to Model[Sample, StockSolution, "5% DIEA in DCM"] Deprotonation has been set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Deprotonation
Indicates if an optional deprotonation step is performed between the deprotection and capping steps of synthesis cycle.
DeprotonationVolume
The volume of DeprotonationSolution added to each reaction vessel during an optional deprotonation step of a synthesis cycle.
Default Calculation: Will automatically resolve to 3 Milliliter plus an additonal .2 Milliliter for each 1 Micromole of synthesis Scale, rounded up to the nearest .5 Milliliter.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
DeprotonationTime
The amount of time that each reaction vessel is exposed to the DeprotonationSolution during each deprotonation step of a synthesis cycle.
NumberOfDeprotonations
The number of repetitions of mixing the resin with the DeprotonationSolution during the deprotonation step of a synthesis cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
NumberOfDeprotonationWashes
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
Capping
CappingSolution
The model or sample objects that represents the solution that will be to cap any remaining uncoupled sites from growing further during the synthesis to aid in later purification of the truncations.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
InitialCapping
Default Calculation: Will automatically resolve to True of DownloadResin been set to False or False if DownloadResin has been set to True.
FinalCapping
Indicates if a final capping step will be done as part of the last synthesis cycle before the start of cleavage.
CappingVolume
The volume of CappingSolution added to each reaction vessel during the capping step of a main synthesis cycle.
Default Calculation: Will automatically resolve to 3 Milliliter plus an additonal .2 Milliliter for each Micromole of synthesis Scale rounded up to nearest .5 Milliliter
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters.
CappingTime
The amount of time that each reaction vessel is exposed to the CappingSolution during each capping step of synthesis cycle.
NumberOfCappings
The number of times that each reaction vessel is capped during each capping step of a synthesis cycle.
NumberOfCappingWashes
The number of wash solution washes each reaction vessel undergoes after the capping step of a main synthesis cycle.
Monomer Activation
ActivationSolution
The model or object representing the mix of preactivation and base solutions used to activate the monomers prior to coupling during a synthesis cycle.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Monomers
Default Calculation: Automatic will resolved to default options for all monomers needed for the synthesis.
Programmatic Pattern: {{SequenceP, ObjectP[{Model[Sample], Object[Sample]}] | _String}..} | Automatic
RecoupMonomers
Indicates if any left over monomer solutions will be stored or discarded at the conclusion of a synthesis.
MonomerPreactivation
Determines if the monomer will be preactivated directly on the resin, in a separate reaction vessel or not at all.
Default Calculation: The maximum number of oligomers that can be concurrently synthesis depends on this option. Each ex situ preactivation requires an additional position on the instrument that could be otherwise used for synthesis when no preactivation or in situ preactivation is used.
ActivationVolume
Default Calculation: Will resolve automatically to .2 Milliliter per Micromole of synthesis scale Scale, rounded up to the nearest .5 Milliliter.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters.
ActivationTime
MonomerVolume
The volume of monomer solution added for each reaction vessel to the preactivation vessel for a micromole scale synthesis.
Default Calculation: Will resolve automatically to .2 Milliliter per Micromole of synthesis Scale, rounded up to the nearest .5 Milliliter.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters.
Download Monomer Activation
DownloadActivationSolution
Default Calculation: Will automatically resolve to value specified by ActivationSolution if DownloadResin is set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
DownloadActivationVolume
The volume of activator solution added to each preactivation vessel when preactivation monomer for coupling to undownloaded resin.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
DownloadActivationTime
The amount of time for which the monomer solution will be mixed with the activation solution prior to a resin download cycle.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 30 minutes or Null.
DownloadMonomer
Default Calculation: Will automatically resolve to a default monomer solution for the monomer being used to download the resin.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
DownloadMonomerVolume
Default Calculation: Will automatically resolve to 1 Milliliter. The entire volume of a DownloadMonomer is delivered in a single shot and can be any amount within the accepted range.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
SampleStorage
StorageBuffer
Default Calculation: Will automatically resolve to Model[Sample,"Dimethylformamide, Reagent Grade"] if Cleavage is set to False.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
ResuspensionBuffer
The sample or model object that represents the object that will be used to resuspend the peptide strands after cleavage from the resin.
Default Calculation: Will automatically resolve to Model[Sample,"Milli-Q water"] if Cleavage is set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
StorageVolume
Default Calculation: Automatically resolved to Null if all of the resins are cleaved or to 15 Milliliter/Gram with a 5 Milliliter minimum.
Pattern Description: Greater than or equal to 0.5 milliliters and less than or equal to 25000 milliliters or Null.
ResuspensionVolume
The volume of resuspension buffer that will be used to resuspend the peptide strands after cleavage from the resin.
Pattern Description: Greater than or equal to 0 milliliters and less than or equal to 3 milliliters or Null.
NumberOfResuspensionMixes
Pattern Description: Greater than or equal to 1 and less than or equal to 30 in increments of 1 or Null.
ResuspensionMixTime
The length of time for which the pelleted strands will be mixed in resuspension buffer (by sonication or vortexing).
ResuspensionMixType
ResuspensionMixPrimitives
A complete list of all incubate primitives used to resuspend and mix the strands after trituration. Cannot be provided if the options NumberOfResuspensionMixes,ResuspensionMixTime, and/or ResuspensionMixType are informed.
Cleavage
PrimaryResinShrinkSolution
The model or sample object that represents the primary shrink solution used to wash and dry the resin after last coupling step.
Default Calculation: Will automatically resolve to Model[Sample, "Methanol"] if PrimaryResinShrink is set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
SecondaryResinShrinkSolution
The model or sample object that represents the secondary shrink solution used to wash and dry the resin after last coupling step.
Default Calculation: Will automatically resolve to Model[Sample, "Isopropanol"] if SecondaryResinShrink is set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
CleavageSolution
The sample or model object that represents the cleavage cocktail that will be used to cleave the peptide strands from the resin.
Default Calculation: Will automatically resolve to Model[Sample,StockSolution,"95%TFA-TIPS-H2O"] if Cleavage is set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Cleavage
Indicates if the oligomers will be cleaved from the resin at the end of the synthesis using CleavageSolution. Uncleaved strands will be stored as a resin slurry in StorageBuffer.
PrimaryResinShrink
PrimaryResinShrinkVolume
The volume of primary shrink solution that will be used to wash the resin at the end of the last synthesis cycle.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
PrimaryResinShrinkTime
SecondaryResinShrink
Indicates if the resin is shrunk with secondary shrink solution prior to strand cleavage or storage.
SecondaryResinShrinkVolume
The volume of secondary shrink solution that will be used to wash the resin at the end of the last synthesis cycle.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
SecondaryResinShrinkTime
CleavageVolume
Pattern Description: Greater than or equal to 2.5 milliliters and less than or equal to 25 milliliters or Null.
CleavageTime
NumberOfCleavageCycles
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
Trituration
TriturationSolution
The sample or model object that represents the solution that will be used to triturate the peptide strands after cleavage from the resin.
Default Calculation: Will automatically resolve to Model[Sample,"Diethyl ether"] if Cleavage is set to True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
TriturationVolume
The volume of ether that will be used to triturate the peptide strands after cleavage from the resin.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 35 milliliters or Null.
NumberOfTriturationCycles
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
TriturationTime
TriturationTemperature
Download Deprotection
DownloadDeprotectionVolume
Default Calculation: Automatic will resolve to 3 Milliliter plus an additonal .5 Milliliter for each 2.5 umol scale for PeptideSynthesizer syntheses.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
DownloadDeprotectionTime
The amount of time that each reaction vessel is exposed to the deprotection solution during each deprotection step of a resin download cycle.
Default Calculation: Will automatically resolve to the same value as DeprotectionTime is DownloadResin is set to True.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 30 minutes or Null.
NumberOfDownloadDeprotections
The number of times that each reaction vessel will be deprotected per deprotection step of a synthesis cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
NumberOfDownloadDeprotectionWashes
The number of wash solution washes each reaction vessel undergoes after the deprotection step of the synthesis cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
DownloadDeprotonationVolume
The volume of DeprotonationSolution added to each reaction vessel during a deprotonation step of a resin download cycle.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
DownloadDeprotonationTime
The amount of time for which the resin will be mixed with DeprotonationSolution for each deprotonation step of a resin download cycle.
Pattern Description: Greater than or equal to 1 minute and less than or equal to 30 minutes or Null.
NumberOfDownloadDeprotonations
The number of repetitions of mixing the resin with the DeprotonationSolution during a resin download cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
NumberOfDownloadDeprotonationWashes
The number of washes each reaction vessel will undergo after a deprotonation step during a resin download cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
Download Capping
DownloadCappingVolume
The volume of CappingSolution added to each reaction vessel during the capping step of of a resin download cycle.
Default Calculation: Will automatically resolve to 3 Milliliter plus an additonal .2 Milliliter per each umol of Scale.
Pattern Description: Greater than or equal to 1 milliliter and less than or equal to 25 milliliters or Null.
DownloadCappingTime
The amount of time for which the resin will be shaken with CappingSolution per capping step of a resin download cycle.
NumberOfDownloadCappings
The number of times that each reaction vessel is capped during each capping step of a resin download cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
NumberOfDownloadCappingWashes
The number of wash solution washes each reaction vessel undergoes after the capping step of a resin download cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
Coupling
CouplingTime
The amount of time that each reaction vessel is exposed to the coupling solution during each coupling step of a synthesis cycle.
NumberOfCouplingWashes
The number of wash solution washes each reaction vessel undergoes after the coupling step of a synthesis cycle.
DoubleCoupling
Specifies the cycle at which monomers are double coupled. All couplings will be performed twice following the cycle number specified.
Download Coupling
DownloadCouplingTime
The amount of time for which the resin mixed be mixed with coupling solution during a coupling step of a resin download cycle.
NumberOfDownloadCouplingWashes
The number of wash solution washes each reaction vessel undergoes after the coupling step of a resin download cycle.
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
Post Experiment
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
Example Calls
Resin
The resin that will be used for the synthesis can be controlled via the Resin option. If you would like to use a resin that is pre-downloaded with a monomer, indicate that via the option DownloadResin->False and specify which Resin to use. For example, here we perform one synthesis with a regular resin (where the first monomer will be downloaded onto the resin), and another synthesis with a resin that already contains the first monomer:

Cleavage and Sample Storage
By default, the synthesized strands are cleaved from the resin at the end of the synthesis and then resuspended in distilled water. If desired, it is possible to keep the strands on the resin by specifying Cleavage -> False, in which case the uncleaved strands will be stored as a resin slurry in storage buffer. The parameters for these manipulations (such as the buffer and buffer amount) are controlled via the set of Resuspension and Storage options, respectively. In the following example, the first synthesized peptide is being cleaved in 2.5 mL of cleavage solution and then resuspended in 1 mL of resuspension solution, while the second strand is kept uncleaved and is stored as a slurry in 2 mL of storage solution:

ExperimentPeptideSynthesis offers flexibility in the way that the cleaved and dried peptide pellet is resuspended in the final resuspension solution. The type of mixing can be specified by ResuspensionMixType as either Pipette, Sonicate or Vortex. NumberOfResuspensionMixes and ResuspensionMixTime provide control over the duration of the mixing. If more thorough mixing is desired, sequential mixing steps can be provided by a list of instruction primitives for each strand. In the following example, the peptide pellet is first resuspended by pipetting up and down 10 times, followed by a 5 minutes sonication and vortexing step to ensure full solubilization of the peptides

Warnings and Errors
Messages (114)
DownloadedResinNeeded (1)
IncompatibleResuspensionMixPrimitives (1)
InsufficientSolventVolume (1)
MismatchedResinAndStrand (1)
NumberOfInputs (1)
RequiredOption (1)
ResinNeeded (1)
TooManyMonomers (1)
UnknownDownloadMonomer (1)
UnneededCleavageOptions (3)
UnneededDeprotonationOptions (1)
UnneededDownloadResinOptions (1)
UnneededResuspensionOptions (1)
UnneededShrinkOptions (1)
UnneededStorageOptions (1)
UnneededSwellOptions (1)
WrongResin (1)
CleavageSolution (2)
CleavageTime (2)
CleavageVolume (2)
DeprotonationSolution (3)
DeprotonationTime (2)
DeprotonationVolume (2)
DownloadActivationSolution (2)
DownloadActivationTime (2)
DownloadActivationVolume (2)
DownloadCappingTime (2)
DownloadCappingVolume (2)
DownloadCouplingTime (2)
DownloadDeprotectionTime (2)
DownloadDeprotectionVolume (2)
DownloadDeprotonationTime (2)
DownloadDeprotonationVolume (2)
DownloadedResinNeeded (1)
DownloadMonomer (2)
DownloadMonomerVolume (1)
MismatchedResinAndStrand (1)
NumberOfCleavageCycles (2)
NumberOfDeprotonations (2)
NumberOfDeprotonationWashes (2)
NumberOfDownloadCappings (2)
NumberOfDownloadCappingWashes (2)
NumberOfDownloadCouplingWashes (2)
NumberOfDownloadDeprotections (2)
NumberOfDownloadDeprotectionWashes (2)
NumberOfDownloadDeprotonations (2)
NumberOfDownloadDeprotonationWashes (2)
NumberOfSwellCycles (1)
NumberOfTriturationCycles (2)
PrimaryResinShrinkSolution (2)
PrimaryResinShrinkTime (2)
PrimaryResinShrinkVolume (2)
ResinNeeded (1)
ResuspensionBuffer (2)
ResuspensionVolume (2)
SecondaryResinShrinkSolution (2)
SecondaryResinShrinkTime (2)
SecondaryResinShrinkVolume (2)
StorageBuffer (2)
StorageVolume (2)
TriturationSolution (2)
TriturationTemperature (2)
TriturationTime (2)
TriturationVolume (2)
Possible Issues
Last modified on Wed 2 Aug 2023 10:30:50