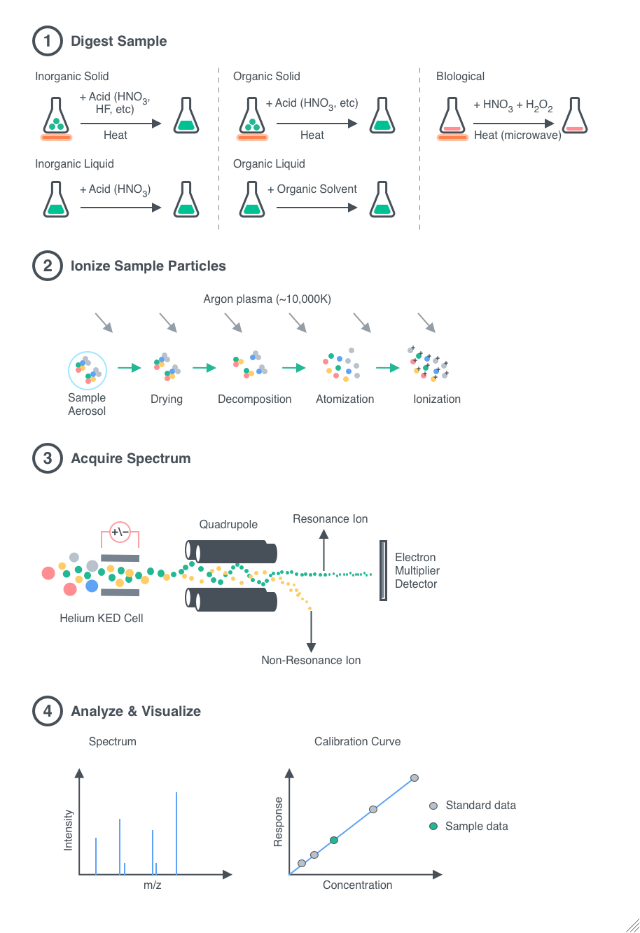
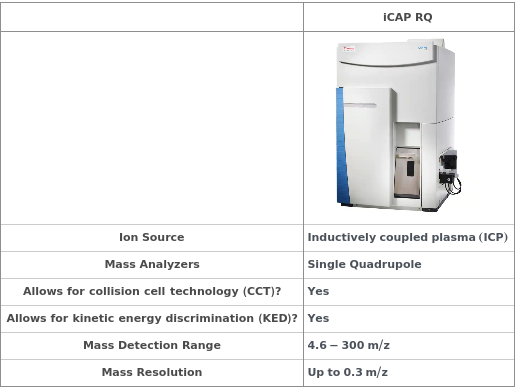
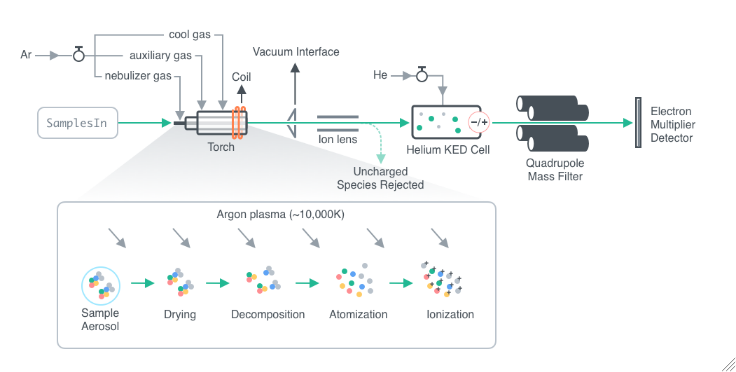
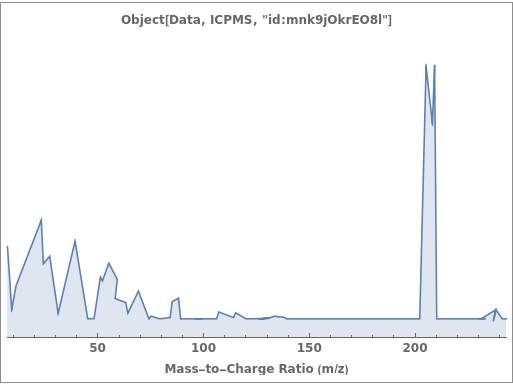
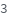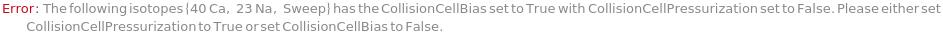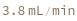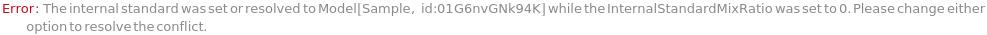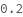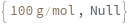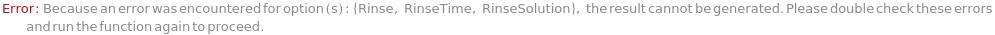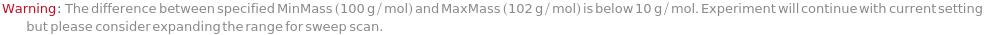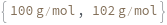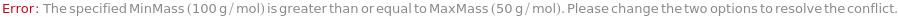General
Instrument
Instrument which is used to atomize, ionize and analyze the elemental composition of the input analyte.
Default Value: Model[Instrument, MassSpectrometer, iCAP RQ ICP-MS]
Pattern Description: An object of type or subtype Model[Instrument, MassSpectrometer] or Object[Instrument, MassSpectrometer]
Programmatic Pattern: ObjectP[{Model[Instrument, MassSpectrometer], Object[Instrument, MassSpectrometer]}]
NumberOfReplicates
Number of times each of the input samples should be analyzed using identical experimental parameters.
Pattern Description: Greater than or equal to 2 in increments of 1 or Null.
Programmatic Pattern: GreaterEqualP[2, 1] | Null
Standards
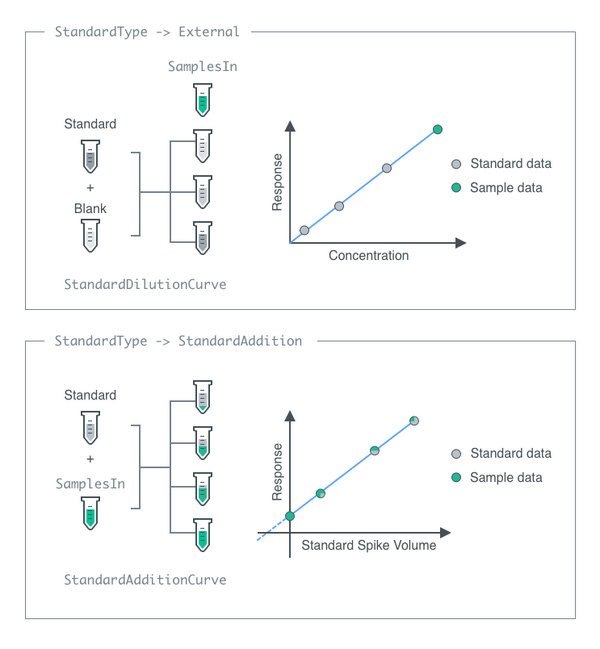
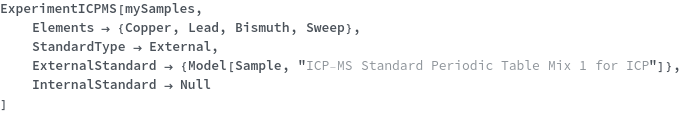
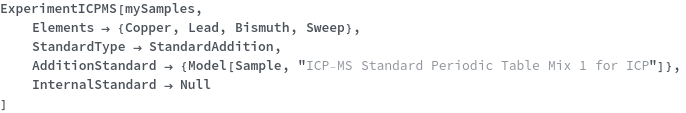
StandardType
Select whether external standard, standard addition or no standard will be used for quantification. StandardAddition are known concentrations of analytes at a given mass that are mixed into the sample prior to introduction into the instrument. External standards are secondary samples of known analytes and concentrations that are injected and measured separately with the instrument.
Default Calculation: Automatically set to External if any of ExternalStandard, ExternalStandardElement or StandardDilutionCurve is specified; otherwise, automatically set to StandardAddition if any of AdditionStandard, StandardAdditionElements, StandardAdditionCurve, StandardSpiledSample is specified; otherwise if QuantifyConcentration for any Elements is set to True, set to External. If all above doesn't hold, set to None.
Pattern Description: External, StandardAddition, or None.
Programmatic Pattern: (External | StandardAddition | None) | Automatic
ExternalStandardElements
Nuclei or element present in the standard solution with known concentrations before dilution.
Default Calculation: Automatically set to all elements present in the input standard's Composition field. If there are no elements in the Composition and this option is unspecified, an error is thrown.
Pattern Description: Manually select all elements and automatically reads concentration or Manually select all elements and enter concentration or Manually select all isotopes and enter concentration or Null.
Programmatic Pattern: ((DuplicateFreeListableP[ICPMSElementP] | {({ICPMSElementP, GreaterEqualP[0*(Milligram/Liter)]} | {ICPMSElementP, GreaterEqualP[0*Micromolar]})..} | {({ICPMSNucleusP, GreaterEqualP[0*(Milligram/Liter)]} | {ICPMSNucleusP, GreaterEqualP[0*Micromolar]})..}) | Automatic) | Null
Index Matches to: ExternalStandard
Digestion
Digestion
Indicates if microwave digestion is performed on the samples in order to fully dissolve the elements into the liquid matrix before injecting into the mass spectrometer.
Default Calculation: Is automatically set to False for acidic aqueous samples and liquid organic non-biological samples, otherwise set to True.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: experiment samples
SampleAmount
The amount of sample that is loaded into the ICPMS instrument for measurement, after all sample preparation procedures but before adding any InternalStandard and/or AdditionStandard.
Default Value: 3000 microliters
Default Calculation: The default sample amount is set to 3000 Microliter.
Pattern Description: Greater than or equal to 2000 microliters and less than or equal to 5000 microliters.
Programmatic Pattern: RangeP[2000*Microliter, 5000*Microliter]
Index Matches to: experiment samples
DigestionInstrument
The reactor used to perform the microwave digestion.
Default Value: Model[Instrument, Reactor, Microwave, Discover SP-D 80]
Pattern Description: An object of type or subtype Object[Instrument, Reactor, Microwave] or Model[Instrument, Reactor, Microwave] or Null.
Programmatic Pattern: ObjectP[{Object[Instrument, Reactor, Microwave], Model[Instrument, Reactor, Microwave]}] | Null
SampleType
Specifies if the sample is primarily composed of organic material (solid or liquid), inorganic material (solid or liquid), is a tablet formulation, or is biological in origin, such as tissue culture or cell culture sample.
Default Calculation: The sample type is set to Organic for any solid and liquid samples by default, and Tablet for samples with Tablet -> True.
Pattern Description: Organic, Inorganic, Tablet, or Biological or Null.
Programmatic Pattern: ((Organic | Inorganic | Tablet | Biological) | Automatic) | Null
Index Matches to: experiment samples
SampleDigestionAmount
The amount of sample that is mixed with DigestionAgents to fully solublize any solid components.
Default Calculation: The default sample amount is set to 10 Milligram or 100 Microliter for solid and liquids samples, respectively.
Pattern Description: Mass or Volume or Null.
Programmatic Pattern: ((RangeP[1*Microliter, 20000*Microliter] | RangeP[1*Milligram, 500*Milligram]) | Automatic) | Null
Index Matches to: experiment samples
PreparedPreDigestionSample
Indicates if the member of SampleIn is already mixed with an appropriate digestion agent. Setting PreparedSample -> True will change the upper limit on the SampleAmount to 20 mL, allowing it to occupy the full volume of the microwave vessel.
Default Calculation: Automatically set to True for liquid samples which contain greater than 50 % of a standard digestion agent.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
DigestionAgents
The sample model and volume of chemical digestion agents used to digest and dissolve the input sample in the form of {Model[Sample], volume}.
Default Calculation: Automatically set to Nitric acid : H2O2 = 9 : 1 for organic samples, Nitric acid : HCl = 9 : 1 for inorganic samples, and Nitric acid : H2O2 : HCl = 8 : 1 : 1 for tablet samples.
Pattern Description: List of one or more {Digestion Agent, Volume} entries or Null.
Programmatic Pattern: ((Null | {{Model[Sample, "Nitric Acid 70% (TraceMetal Grade)"] | Model[Sample, "Hydrochloric Acid 37%, (TraceMetal Grade)"] | Model[Sample, "Sulfuric Acid 96% (TraceMetal Grade)"] | Model[Sample, "Phosphoric Acid 85% (>99.999% trace metals basis)"] | Model[Sample, "Hydrogen Peroxide 30% for ultratrace analysis"] | Model[Sample, "Milli-Q water"] | Model[Sample, "id:qdkmxzqpeaRY"] | Model[Sample, "id:N80DNj1WpbLD"] | Model[Sample, "id:n0k9mG8Z1b4p"] | Model[Sample, "id:lYq9jRx5nvmO"] | Model[Sample, "id:xRO9n3BGjKb5"] | Model[Sample, "id:8qZ1VWNmdLBD"], RangeP[0*Milliliter, 20*Milliliter]}..}) | Automatic) | Null
Index Matches to: experiment samples
DigestionTemperature
The temperature to which the sample is heated for the duration of the DigestionDuration.
Default Calculation: Automatically set to 200 Celsius when DigestionTemperatureProfile is not specified.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 300 degrees Celsius or Null.
Programmatic Pattern: ((Null | RangeP[25*Celsius, 300*Celsius]) | Automatic) | Null
Index Matches to: experiment samples
DigestionDuration
The amount of time for which the sample is incubated at the set DigestionTemperature during digestion.
Default Calculation: When DigestionTemperatureProfile is specified, DigestionDuration matches the length of the final isothermal segment. Otherwise it is set to 10 minutes.
Pattern Description: Greater than or equal to 0 hours and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Hour, Min[$MaxExperimentTime, 99*Hour]] | Automatic) | Null
Index Matches to: experiment samples
DigestionRampDuration
The amount of time taken for the sample chamber temperature from ambient temperature to reach the DigestionTemperature.
Default Calculation: Is automatically set to heat at a rate of 40 C/minute unless DigestionTemperatureProfile is specified.
Pattern Description: Greater than or equal to 0 hours and less than or equal to 72 hours or Null.
Programmatic Pattern: ((Null | RangeP[0*Hour, Min[$MaxExperimentTime, 99*Hour]]) | Automatic) | Null
Index Matches to: experiment samples
DigestionTemperatureProfile
The heating profile for the reaction mixture in the form {{Time, Target Temperature}..}. Consecutive entries with different temperatures result in a linear ramp between the temperatures, while consecutive entries with the same temperature indicate an isothermal region at the specified temperature.
Default Calculation: When DigestionTemperature, DigestionRampTime, or DigestionTime are specified, this option set to {{DigestionRampDuration, DigestionTemperature},{DigestionRampDuration + DigestionDuration, DigestionTemperature}}.
Pattern Description: List of one or more {Time, Target Temperature} entries or Null.
Programmatic Pattern: ((Null | {{RangeP[0*Hour, Min[$MaxExperimentTime, 99*Hour]], RangeP[25*Celsius, 300*Celsius]}..}) | Automatic) | Null
Index Matches to: experiment samples
DigestionMixRateProfile
The relative rate of the magnetic stir bar rotation that will be used to mix the sample, either for the duration of the digestion (fixed), or from the designated time point to the next time point (variable). For safety reasons, the sample must be mixed during microwave heating.
Pattern Description: List of one or more {Time, Mix Rate} entries or Low, Medium, or High or Null.
Programmatic Pattern: ((Low | Medium | High) | {{RangeP[0*Hour, Min[$MaxExperimentTime, 99*Hour]], Low | Medium | High}..}) | Null
Index Matches to: experiment samples
DigestionMaxPower
The maximum power of the microwave radiation output that will be used during heating.
Default Calculation: Automatically set to 200 Watt if SampleType -> Organic, and automatically set to 300 Watt For SampleType -> Tablet or Inorganic.
Pattern Description: Greater than or equal to 1 watt and less than or equal to 300 watts or Null.
Programmatic Pattern: (RangeP[1*Watt, 300*Watt] | Automatic) | Null
Index Matches to: experiment samples
DigestionMaxPressure
The pressure at which the magnetron will cease to heat the reaction vessel. If the vessel internal pressure exceeds 500 PSI, the instrument will cease heating regardless of this option.
Default Value: 250 pounds
‐force per inch squared
Pattern Description: Greater than or equal to 1 pound
‐force per inch squared and less than or equal to 500 pounds
‐force per inch squared or Null.
Programmatic Pattern: RangeP[1*PSI, 500*PSI] | Null
Index Matches to: experiment samples
DigestionPressureVenting
Indicates if the reaction vessel is vented when pressure rise above DigestionPressureVentingTriggers.
Default Calculation: Automatically set to True for Inorganic and Tablet samples if DigestionPressureVentingTriggers or DigestionTargetPressureReduction is specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
DigestionPressureVentingTriggers
The set point pressures at which venting will begin, and the number of times that the system will vent the vessel in an attempt to reduce the pressure by the value of TargetPressureReduction. If the pressure set points are not reached, no venting will occur. Be aware that excessive venting may result in sample evaporation, leading to dry out of sample, loss of sample and damage of reaction vessel.
Default Calculation: If PressureVenting -> True, the PressureVentingTriggers are set to include a venting step at 50 PSI (2 attempts) with additional venting set according to SampleType. If SampleType -> Organic, additional venting occurs at 25 PSI increments starting at 225 PSI with 2 venting attempts at each pressure point until 350 PSI, for which venting is set to 100 attempts. Inorganic samples utilize additional venting at 400 PSI, using 100 attempts.
Pattern Description: List of one or more {Trigger Pressure, Number of Ventings} entries or Null.
Programmatic Pattern: ((Null | {{RangeP[1*PSI, 400*PSI], RangeP[1, 127]}..}) | Automatic) | Null
Index Matches to: experiment samples
DigestionTargetPressureReduction
The target drop in pressure during PressureVenting. Venting is conducted according to the PressureVentingTriggers.
Default Calculation: When PressureVenting -> True, the TargetPressureReduction is set based on the value of SampleType. Organic samples and tablets require more frequent venting until 25 PSI below DigestionPressureVentingTriggers, while inorganic samples may be vented less frequently until 40 PSI below DigestionPressureVentingTriggers.
Pattern Description: Greater than or equal to 1 pound
‐force per inch squared and less than or equal to 300 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((Null | RangeP[1*PSI, 300*PSI]) | Automatic) | Null
Index Matches to: experiment samples
DigestionOutputAliquot
The maximum amount of the reaction mixture that is aliquoted into the ContainerOut. If the volume of sample after experiment falls below DigestionOutputAliquot, the full fraction will be collected. The remaining reaction mixture is discarded.
Default Value: 3 milliliters
Default Calculation: For ICPMS applications, if StandardType is set to External or None, this option is automatically set to 150 ul. If StandardType is set to Internal, set to 150 ul times the number of internal standard additions specified by the InternalStandardAdditionCurve.
Pattern Description: All or greater than or equal to 0 milliliters and less than or equal to 20 milliliters or Null.
Programmatic Pattern: (Null | (RangeP[0*Milliliter, 20*Milliliter] | All)) | Null
Index Matches to: experiment samples
DiluteDigestionOutputAliquot
Indicates if the OutputAliquot is added to a specified volume (PostDigestionDiluentVolume) of Diluent prior to storage or use in subsequent experiments. Dilution reduces the risk and cost associated with storage of caustic/oxidizing reagents commonly employed in digestion protocols.
Default Calculation: When Diluent and PostDigestionDiluentVolume are specified, this will automatically set to True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
PostDigestionDiluent
The solution used to dilute the OutputAliquot of the digested sample.
Default Calculation: When DiluteOutputAliquot -> True, the default diluent is automatically set to Model[Sample, "Trace metal grade water"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((Null | (ObjectP[{Model[Sample], Object[Sample]}] | _String)) | Automatic) | Null
Index Matches to: experiment samples
PostDigestionDiluentVolume
The volume of diluent into which the OutputAliquot will be added. User should only specify one of the 3 options: PostDigestionDiluentVolume, PostDigestionDilutionFactor, PostDigestionSampleVolume.
Pattern Description: Greater than or equal to 0 liters and less than or equal to 1 liter or Null.
Programmatic Pattern: (Null | RangeP[0*Liter, 1*Liter]) | Null
Index Matches to: experiment samples
PostDigestionDilutionFactor
The desired dilution factor for this mixture. User should only specify one of the 3 options: PostDigestionDiluentVolume, PostDigestionDilutionFactor, PostDigestionSampleVolume.
Default Calculation: When DiluteOutputAliquot -> True, automatically set to a value so that the volume after dilution is 3 ml.
Pattern Description: Greater than or equal to 1 and less than or equal to 1000 or Null.
Programmatic Pattern: ((Null | RangeP[1, 1000]) | Automatic) | Null
Index Matches to: experiment samples
PostDigestionSampleVolume
The volume of output sample after DigestionOutputAliquot has been removed, and subsequently been diluted by the PostDigestionDiluentVolume of the provided Diluent sample. User should only specify one of the 3 options: PostDigestionDiluentVolume, PostDigestionDilutionFactor, PostDigestionSampleVolume.
Default Calculation: Automatically set to equal to the sum of PostDigestionDiluentVolume and DigestionOutputAliquot if PostDigestionDiluentVolume is specified, otherwise automatically set to 3 ml.
Pattern Description: Target Volume or Null.
Programmatic Pattern: (Null | (Alternatives[RangeP[0*Liter, 1*Liter]])) | Null
Index Matches to: experiment samples
DigestionContainerOut
The container into which the OutputAliquotVolume or dilutions thereof is placed as the output of digestion. If StandardType is set to Internal, the sample will be subjected to internal standard addition before injecting into ICPMS instrument, otherwise the sample will be directly injected to ICPMS instrument.
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[Model[Container]] | Automatic) | Null
Index Matches to: experiment samples
Standard
InternalStandard
Standard sample that is added to all samples, including standards, blanks and user-provided samples. InternalStandard contains elements which do not exist in any other samples, and which its detector response is measured along with other Elements to account for matrix effects and instrument drift.
Default Calculation: If there exists any Element with InternalStandardElement set to True, automatically choose a standard that contains these elements but do not contain any other elements in Elements option. If no such element exists, will try to find one standard that do not contain any elements need quantification, and automatically add one element in Elements option as InternalStandardElement.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Automatic or Null.
Programmatic Pattern: (((Automatic | Null) | (ObjectP[{Model[Sample], Object[Sample]}] | _String)) | Automatic) | Null
InternalStandardMixRatio
Volume ratio of InternalStandard to each sample.
Default Calculation: Automatically set to 0.111 if there's any InternalStandard, otherwise set to 0.
Pattern Description: Greater than or equal to 0 and less than or equal to 1.
Programmatic Pattern: RangeP[0, 1] | Automatic
ExternalStandard
Calibration standard for quantification of analyte elements or nuclei.
Default Calculation: Automatically selects one or more standards that contains all values in the Analyte option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
Index Matches to: ExternalStandard
StandardDilutionCurve
The collection of dilutions that are performed on the ExternalStandard prior to injecting the standard samples into the instrument.
Default Calculation: Automatically set to a list of 3 dilution factors such that highest concentration of the ExternalStandardElement becomes 1, 3 and 10 mg/ml. If the maximum concentration is below 10 mg/ml, set the dilution factor to be 1, 3 and 10 with 10 ml total volume after dilution.
Pattern Description: Automatic or Null or Fixed Dilution Factor or Fixed Dilution Volume.
Programmatic Pattern: (((Automatic | Null) | {{GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter]}..} | {{GreaterEqualP[0*Microliter], GreaterEqualP[1]}..}) | Automatic) | Null
Index Matches to: ExternalStandard
StandardAddedSample
Indicate if the input sample has already been spiked with standard sample for concentration measurement via standard addition.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
Index Matches to: experiment samples
AdditionStandard
standard samples being added to the sample for standard addition.
Default Calculation: Automatically selects one standard that contains all Elements analytes in the input sample; if that's impossible, selects one standard that contains the first Elements in the input sample.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
StandardAdditionCurve
The collection of StandardAddition performed on the input sample prior to injecting the standard samples into the instrument. If StandardAddedSample is set to True for any sample, these samples will no longer be mixed with AdditionStandard again, and This option should be set to Null.
Default Calculation: Automatically set to a list of 3 volume ratios such that concentration of the first AdditionStandard's Analyte becomes 1, 3 and 10 mg/ml. If the standard concentration is below 20 mg/ml, will add to consist 20%, 40% and 100% of SampleAmount. If StandardAddedSample set to True, this option automatically set to Null
Pattern Description: Automatic or Null or Fixed ratio or Fixed volume.
Programmatic Pattern: (((Automatic | Null) | {{GreaterEqualP[0]}..} | {{RangeP[0*Milliliter, 3*Milliliter]}..}) | Automatic) | Null
Index Matches to: experiment samples
StandardAdditionElements
Nuclei or element present in the standard solution with known concentrations before dilution. For samples with StandardAddedSample set to True, concentration in this option instead refers to the value after dilution.
Default Calculation: Automatically set to all elements present in the input standard's Composition field if StandardAddedSample is set to False, or to all elements from monoatomic components in the input sample's Composition field if StandardAddedSample is set to True. If there are no elements in the Composition and this option is unspecified, an error is thrown.
Pattern Description: Manually select all elements and automatically reads concentration or Manually select all elements and enter concentration or Null.
Programmatic Pattern: ((DuplicateFreeListableP[ICPMSElementP] | {({ICPMSElementP, GreaterEqualP[0*(Milligram/Liter)]} | {ICPMSElementP, GreaterEqualP[0*Micromolar]})..}) | Automatic) | Null
Index Matches to: experiment samples
Blank
Blank
Sample containing no analyte used to measure the background levels of any ions in the matrix.
Default Value: Model[Sample, StockSolution, 5% Nitric Acid, trace metal grade]
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Object[Sample], Model[Sample]}] | _String
Flushing
Rinse
Determine whether rinse of inlet line is performed after a set number of samples.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
RinseSolution
Solution for the autosampler to draw between each run to flush the system.
Default Calculation: Is automatically set to match with Blank if Rinse is set to True.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
RinseTime
Duration of time for each rinse.
Default Calculation: Is automatically set to 60 seconds if Rinse is set to True.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 180 seconds or Null.
Programmatic Pattern: (RangeP[0*Second, 180*Second] | Automatic) | Null
Sample Loading
InjectionDuration
Duration of the input sample being continuously injected into the instrument for each measurement.
Default Value: 30 seconds
Default Calculation: Automatically set to 30 seconds.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 120 seconds.
Programmatic Pattern: RangeP[0*Second, 120*Second]
Analysis
ConeDiameter
Indicates diameter of the opening of vacuum interface cone. High cone diameter will allow more plasma into the vacuum chamber for analysis. For higher concentration analyte low cone diameter should be used to block more interferences to minimize noise, while for lower concentration analyte high cone diameter should be used to allow more plasma in to maximize signal.
Default Value: 3.5 millimeters
Pattern Description: 2.8 millimeters, 3.5 millimeters, or 4.5 millimeters.
Programmatic Pattern: 2.8*Millimeter | 3.5*Millimeter | 4.5*Millimeter
IsotopeAbundanceThreshold
Select threshold so that isotopes whose abundance is above this value will be selected for detection besides the most abundant one. This option is only valid when Elements -> Automatic or a list of elements, will be ignored when list of isotopes is provided as entry for Elements option.
Default Value: 20 percent
Pattern Description: Greater than or equal to 0 percent and less than or equal to 50 percent.
Programmatic Pattern: RangeP[0*Percent, 50*Percent]
QuantifyElements
Select if concentration of elements will be quantified in addition to each isotopes.
Pattern Description: True or False.
Programmatic Pattern: BooleanP
Elements
Nuclei or element to be measured by tuning the quadrupole to select the mass of the element that matches the atomic weight. When elements are selected, only the most abundant isotope and isotopes above IsotopeAbundanceThreshold (default 20%) will be quantified. If Elements is set to Sweep then a full spectrum will be acquired. Should also include elements from InternalStandard.
Default Calculation: Automatically set to elements matching the atomic metals that present in the molecules of the sample composition and InternalStandard composition plus Sweep. If there are no metallic elements in the Composition and this option is unspecified, automatically set to Sweep.
Pattern Description: Elements (Most Abundant Isotope) or Isotopes.
Programmatic Pattern: (ICPMSElementP | ICPMSNucleusP) | Automatic
Index Matches to: Elements
DwellTime
Time spend to measure a single analyte of a single sample.
Default Calculation: Automatically set to 1 ms for Sweep and 10 ms for any other Elements
Pattern Description: Greater than or equal to 0.1 milliseconds and less than or equal to 5000 milliseconds.
Programmatic Pattern: RangeP[0.1*Millisecond, 5000*Millisecond] | Automatic
Index Matches to: Elements
InternalStandardElement
Indicates if the particular element comes from InternalStandard, therefore should be measured but not quantified.
Default Calculation: Automatically set to False if InternalStandard is set to Null, otherwise will automatically read composition of InternalStandard and set to True for any elements that exist in the InternalStandard.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: Elements
QuantifyConcentration
Indicates if concentrations of the particular element in all input samples are quantified.
Default Calculation: Is automatically set to False when InternalStandardElement set to True, and automatically set to True when StandardType set to anything other than None, and such element can be found in at least one standard.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: Elements
MinMass
When Elements includes Sweep, sets the lower limit of the m/z value for the quadrupole mass scanning range.
Default Calculation: Automatically set to 4.6 g/mol if Elements is set to Sweep.
Pattern Description: Greater than or equal to 1. grams per mole and less than or equal to 300. grams per mole or Null.
Programmatic Pattern: (RangeP[1.*(Gram/Mole), 300.*(Gram/Mole)] | Automatic) | Null
MaxMass
When Elements is set to Sweep, sets the upper limit of the m/z value for the quadrupole mass scanning range.
Default Calculation: Automatically set to 245 g/mol if Elements is set to Sweep.
Pattern Description: Greater than or equal to 1. grams per mole and less than or equal to 300. grams per mole or Null.
Programmatic Pattern: (RangeP[1.*(Gram/Mole), 300.*(Gram/Mole)] | Automatic) | Null
Ionization
PlasmaPower
Electric power of the coil generating the Ar-ion plasma flowing through the torch.
Default Calculation: Is automatically set to Low for light elements and Normal for other elements.
Pattern Description: Low or Normal.
Programmatic Pattern: (Low | Normal) | Automatic
Index Matches to: Elements
Data Acquisition
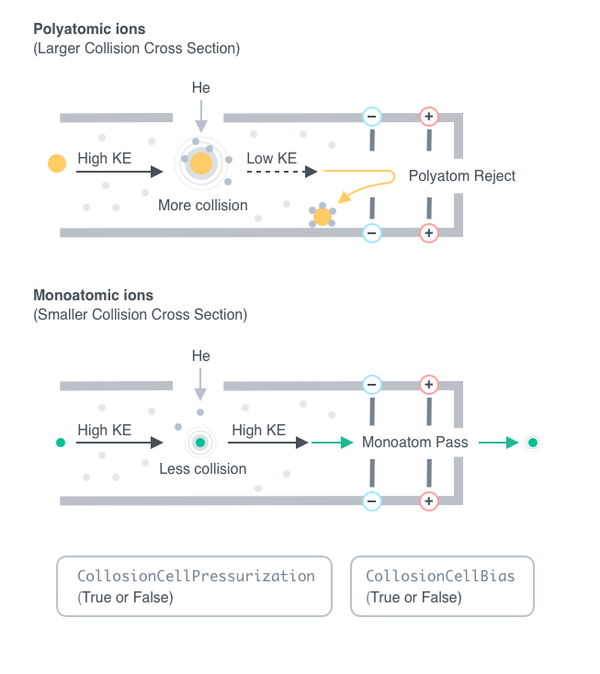
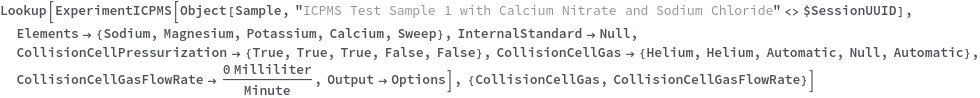
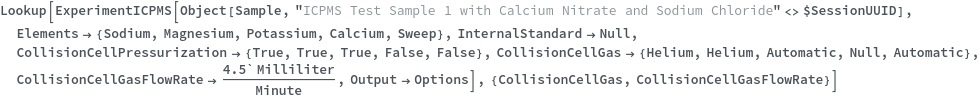
CollisionCellPressurization
Indicate if the collision cell should be pressurized. If the collision cell is pressurized, gas inside will collide with the analyte ions as the ion pass through it. Since polyatomic ions are larger than monoatomic ions they are more likely to collide with the gas, and therefore been blocked from entry of the quadrupole. See Figure 3.2 for more details.
Default Calculation: Is automatically set to True if CollisionCellGas is set to non-Null, or if CollisionCellGasFlowRate is set to non-zero.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: Elements
CollisionCellGas
Indicate which gas type collision cell should be pressurized.
Default Calculation: Is automatically set to Helium if CollisionCellPressurization is set to True.
Pattern Description: Helium or Oxygen or Null.
Programmatic Pattern: (ICPMSCollisionCellGasP | Automatic) | Null
Index Matches to: Elements
CollisionCellGasFlowRate
Indicates the flow rate of gas through collision cell. Refer to Figure X
Default Calculation: Is automatically set to 4.5 ml/min if CollisionCellPressurization is set to True.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 10 milliliters per minute.
Programmatic Pattern: RangeP[(0*Milliliter)/Minute, (10*Milliliter)/Minute] | Automatic
Index Matches to: Elements
CollisionCellBias
Indicates if bias voltage for collision cell is turned on. Refer to Figure X
Default Calculation: Is automatically set to True if CollisionCellPressurization was set to True.
Pattern Description: True or False.
Programmatic Pattern: BooleanP | Automatic
Index Matches to: Elements
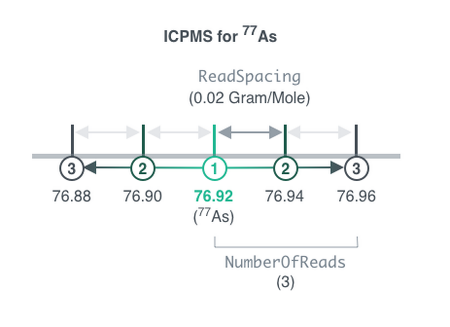
NumberOfReads
Number of redundant readings by the instrument on each side the target mass, see Figure 3.3. This option is not valid for Sweep.
Pattern Description: Greater than or equal to 1 and less than or equal to 13 in increments of 1.
Programmatic Pattern: RangeP[1, 13, 1]
Index Matches to: Elements
ReadSpacing
Distance of mass units between adjacent redundant readings. For Sweep, the minimum allowed value is 0.05 amu. See Figure X2.
Default Calculation: Automatically set to 0.1 Gram/Mole for all Elements except Sweep, and set to 0.2 Gram/Mole for Sweep.
Pattern Description: Greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole.
Programmatic Pattern: RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | Automatic
Index Matches to: Elements
Bandpass
Select the bandwidth of allowed m/z ratio for each reading by the quadrupole. Normal is 0.75 amu at 10% peak height, and High is 0.3 amu at 10% peak height. Set Resolution to High can better differentiate some isotopes, with a price of reduced signal level.
Pattern Description: Normal or High.
Programmatic Pattern: Normal | High
Index Matches to: Elements
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, BacteriaIncubation, MammalianIncubation, TissueCultureCellsIncubation, MicrobialCellsIncubation, MicrobialCellsShakingIncubation, YeastCellsIncubation, YeastCellsShakingIncubation, ViralIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples
Sample Preparation
CrushSample
Indicates if the tablet is crushed to a powder prior to mixing with DigestionAgents.
Default Calculation: Automatically set to True if the SampleType is set to Tablet.
Pattern Description: True or False or Null.
Programmatic Pattern: ((Null | BooleanP) | Automatic) | Null
Index Matches to: experiment samples
PreDigestionMix
Indicates if the reaction mixture is stirred at ambient temperature directly prior to being subjected to microwave heating. Pre-mixing can ensure that a sample is fully dissolved or suspended prior to heating.
Default Calculation: Automatically set to True unless the PreDigestionMixTime and PreDigestionMixRate are not specified.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
PreDigestionMixTime
The amount of time for which the reaction mixture is stirred at ambient temperature directly prior to being subjected to microwave heating.
Default Calculation: Automatically set to 2 Minutes when PreDigestionMix is True.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 2 minutes or Null.
Programmatic Pattern: ((Null | RangeP[0*Minute, 2*Minute]) | Automatic) | Null
Index Matches to: experiment samples
PreDigestionMixRate
The rate at which the reaction mixture is stirred at ambient temperature directly prior to being subjected to microwave heating.
Default Calculation: Automatically set to Medium when PreDigestionMix is True.
Pattern Description: None, Low, Medium, or High or Null.
Programmatic Pattern: ((Null | (None | Low | Medium | High)) | Automatic) | Null
Index Matches to: experiment samples