ExperimentPickColonies
ExperimentPickColonies[Objects]⟹Protocol
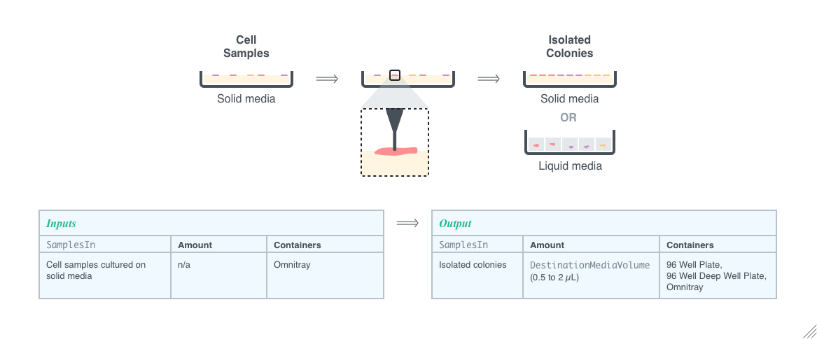
creates a Protocol to pick colonies from the provided sample or container Objects and deposit them into a destination container.
Pick Colonies allows for the isolation of microbial colonies growing on top of a solid medium to duplicate them for further use. The colonies are picked with a thin needle and then transferred to a desired media. The picked colonies can either be placed onto solid or liquid media and then grown and use for further analysis.
Experimental Principles
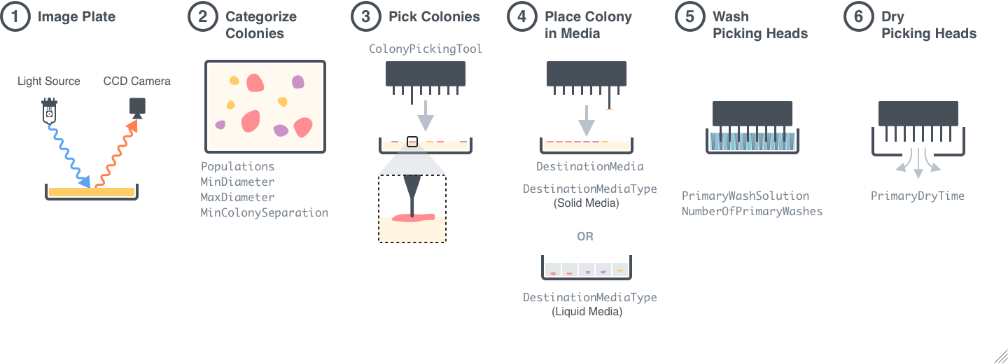
Figure 1.1: Procedural Overview of a PickColonies experiment using a QPix 420 HT Colony Picker. Step 1: Samples are imaged using the CCD camera on the instrument. Step 2: The images are analyzed by the function AnalyzeColonies in order to find the locations of the colonies on the plates. Step 3: The selected colonies are picked by the QPix 420 HT colony picking tool. Step 4: The picked colonies are placed on the desired media. Step 5: The colony picking tool is washed in sanitization solution. Step 6: The colony picking tool is dried by a fan.
Instrumentation
QPix 420 HT

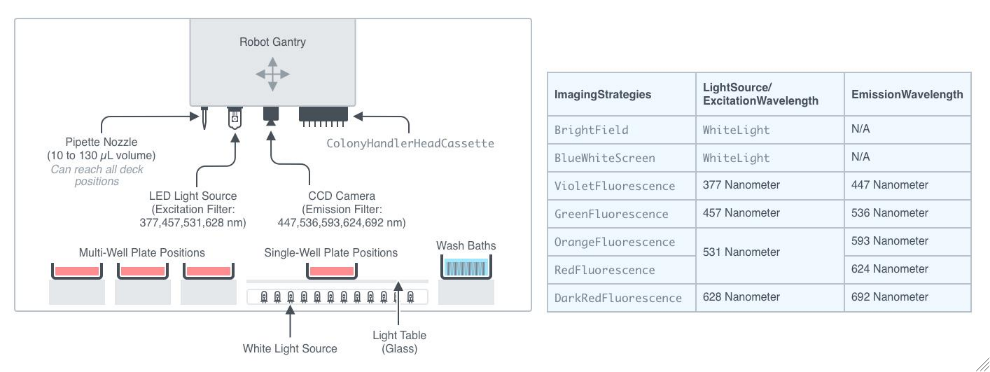
Figure 2.1.1: Overview of the ColonyHandler WorkCell. This figure provides a comprehensive view of the ColonyHandler, showcasing its layout and main components. The QPix instrument is designed to perform multiple tasks including imaging solid media plates, picking colonies from solid media, and plating suspended colonies onto solid media. Its bed features a deck with designated positions for multi-well and single-well SBS plates, a light table, and wash baths for cleaning the pins before and during the process. Above the deck, a robotic gantry moves and is equipped with several components: a pipette nozzle, an LED-based light source, preset filter pairs for fluorescence excitation and emission wavelengths, a monochrome CCD camera, and a position to accept a ColonyHandlerHeadCassette. The robotic gantry moves seamlessly between tasks, and have the capacity to capture multiple types of images.
Diameter
diameterRules
Characterization
Select
The method used to group colonies into a population. Colonies are first ordered by their Diameter, defined as the diameter of a colony is the diameter of a disk with the same area as the colony. If the Select option is Min or Max, the ordered colonies are divided into partitions based on the NumberOfDivisions. From there, the smallest or largest partition is grouped into a population depending on if the Select option is Min or Max. For example, if the Select option is set to Max and the NumberOfDivisions is 10, the colonies will be divided into 10 partitions sorted from smallest to largest and the partition with the largest colonies will be selected as the population. Alternatively, if the Select option is AboveThreshold or BelowThreshold, an absolute cutoff is used to group ordered colonies into a population. The threshold is set by the ThresholdDiameter option. For example, if the Select option is BelowThreshold and the ThresholdDiameter is 0.8 Millimeter, all colonies smaller than 0.8 Millimeter in Diameter are grouped into a population.
ThresholdDiameter
The min or max possible diameter for a colony to be included in the population. When the Select option is AboveThreshold or BelowThreshold, all colonies with a diameter larger or smaller than the ThresholdDiameter, respectively, are grouped into a population. The colony diameter is defined as the diameter of a disk with the same area as the colony.
Pattern Description: Greater than or equal to 0.2 millimeters and less than or equal to 10 millimeters or Null.
NumberOfColonies
Default Calculation: If Select is Min or Max, up to 10 colonies are selected, otherwise All colonies are selected.
NumberOfDivisions
The number of partitions to group colonies into. Partitions are ordered by the Unit Operation feature. If the Select option is Min or Max, the lowest or highest partition will be used, respectively.
PopulationName
Default Calculation: By default, each population is labeled as ColonySelection with sequential integers.
Include
Exclude
Isolation
isolationRules
Characterization
Select
The method used to group colonies into a population. Colonies are first ordered by their Isolation, the shortest distance from the boundary of the colony to the boundary of another. If the Select option is Min or Max, the ordered colonies are divided into partitions based on the NumberOfDivisions. From there, the least isolated or most isolated partition is grouped into a population depending on if the Select option is Min or Max. For example, if the Select option is set to Max and the NumberOfDivisions is 10, the colonies will be divided into 10 partitions sorted from least isolated to most isolated and the partition with the most isolated colonies will be selected as the population. Alternatively, if the Select option is AboveThreshold or BelowThreshold, an absolute cutoff is used to group ordered colonies into a population. The threshold is set by the ThresholdDistance option. For example, if the Select option is AboveThreshold and the ThresholdDistance is 1 Millimeter, all colonies with separation greater than 1 Millimeter are grouped into a population.
ThresholdDistance
The min or max possible isolation for a colony to be included in the population. When the Select option is AboveThreshold or BelowThreshold, all colonies with an isolation distance larger or smaller than the ThresholdDistance, respectively, are grouped into a population. The isolation is the shortest distance from the boundary of the colony to the boundary of another.
NumberOfColonies
Default Calculation: If Select is Min or Max, up to 10 colonies are selected, otherwise All colonies are selected.
NumberOfDivisions
The number of partitions to group colonies into. Partitions are ordered by the Unit Operation feature. If the Select option is Min or Max, the lowest or highest partition will be used, respectively.
PopulationName
Default Calculation: By default, each population is labeled as ColonySelection with sequential integers.
Include
Exclude
Circularity
circularityRules
Characterization
Select
The method used to group colonies into a population. Colonies are first ordered by their Circularity, the ratio of the minor axis to the major axis of the best fit ellipse. If the Select option is Min or Max, the ordered colonies are divided into partitions based on the NumberOfDivisions. From there, the least circular or most circular partition is grouped into a population depending on if the Select option is Min or Max. For example, if the Select option is set to Max and the NumberOfDivisions is 10, the colonies will be divided into 10 partitions sorted from least circular to most circular and the partition with the most circular colonies will be selected as the population. Alternatively, if the Select option is AboveThreshold or BelowThreshold, an absolute cutoff is used to group ordered colonies into a population. The threshold is set by the ThresholdCircularity option. For example, if the Select option is AboveThreshold and the ThresholdCircularity is 0.9, all colonies with a circularity ratio greater than 0.9 are grouped into a population.
ThresholdCircularity
The min or max possible circularity ratio for a colony to be included in the population. When the Select option is AboveThreshold or BelowThreshold, all colonies with a circularity ratio larger or smaller than the ThresholdCircularity, respectively, are grouped into a population. The colony circularity ratio is the ratio of the minor axis to the major axis of the best fit ellipse.
NumberOfColonies
Default Calculation: If Select is Min or Max, up to 10 colonies are selected, otherwise All colonies are selected.
NumberOfDivisions
The number of partitions to group colonies into. Partitions are ordered by the Unit Operation feature. If the Select option is Min or Max, the lowest or highest partition will be used, respectively.
PopulationName
Default Calculation: By default, each population is labeled as ColonySelection with sequential integers.
Include
Exclude
Regularity
regularityRules
Characterization
Select
The method used to group colonies into a population. Colonies are first ordered by their Regularity, the ratio of the area of the colony to the area of a circle with the same perimeter. If the Select option is Min or Max, the ordered colonies are divided into partitions based on the NumberOfDivisions. From there, the least regular or most regular partition is grouped into a population depending on if the Select option is Min or Max. For example, if the Select option is set to Max and the NumberOfDivisions is 10, the colonies will be divided into 10 partitions sorted from least regular to most regular and the partition with the most regular colonies will be selected as the population. Alternatively, if the Select option is AboveThreshold or BelowThreshold, an absolute cutoff is used to group ordered colonies into a population. The threshold is set by the ThresholdRegularity option. For example, if the Select option is AboveThreshold and the ThresholdRegularity is 0.9, all colonies with a regularity ratio greater than 0.9 are grouped into a population.
ThresholdRegularity
The min or max possible regularity ratio for a colony to be included in the population. When the Select option is AboveThreshold or BelowThreshold, all colonies with a regularity ratio larger or smaller than the ThresholdRegularity, respectively, are grouped into a population. The colony regularity ratio is the ratio of the area of the colony to the area of a circle with the same perimeter.
NumberOfColonies
Default Calculation: If Select is Min or Max, up to 10 colonies are selected, otherwise All colonies are selected.
NumberOfDivisions
The number of partitions to group colonies into. Partitions are ordered by the Unit Operation feature. If the Select option is Min or Max, the lowest or highest partition will be used, respectively.
PopulationName
Default Calculation: By default, each population is labeled as ColonySelection with sequential integers.
Include
Exclude
Fluorescence
fluorescenceRules
Characterization
Dye
The fluorophore used to detect colonies that fluoresce at an EmissionWavelength when exposed to an ExcitationWavelength.
Default Calculation: Automatically set to the fluorophore within the excitation/emission range. For VioletFluorescence, set to DAPI. For GreenFluorescence, set to GFP. For OrangeFluorescence, set to Cy3. For RedFluorescence, set to TxRed. For DarkRedFluorescence, set to Cy5.
Pattern Description: DAPI, UltraViolet, GFP, RGBColor[0, 0, 1], Cy3, TxRed, Cy5, or RGBColor[1, 0, 0]
ExcitationWavelength
The wavelength of light that adds energy to colonies and causes them to fluoresce. Pairs with an EmissionWavelength.
Default Calculation: Automatically set to the excitation wavelength specified in the FluorescentExcitationWavelength field of model cell in CellTypes.
EmissionWavelength
The light that the instrument measures to detect colonies fluorescing at a particular wavelength. Pairs with an ExcitationWavelength.
Default Calculation: Automatically set to the emission wavelength specified in the FluorescentEmissionWavelength field of model cell in CellTypes.
Pattern Description: 447 nanometers, 536 nanometers, 593 nanometers, 624 nanometers, or 692 nanometers.
Programmatic Pattern: (447*Nanometer | 536*Nanometer | 593*Nanometer | 624*Nanometer | 692*Nanometer) | Automatic
Select
The method used to group colonies into a population. Colonies are first ordered by their Fluorescence, the average pixel value inside the colony boundary from an image generated using an excitation wavelength and emission filter. If the Select option is Min or Max, the ordered colonies are divided into partitions based on the NumberOfDivisions. From there, the least fluorescing or most fluorescing partition is grouped into a population depending on if the Select option is Min or Max. For example, if the Select option is set to Max and the NumberOfDivisions is 10, the colonies will be divided into 10 partitions sorted from least fluorescing to most fluorescing and the most fluorescing partition will be selected as the population. Alternatively, if the Select option is Positive or Negative, the ordered colonies are first clustered into two groups based on their Fluorescence above or below background intensity. Then, if the Select option is Positive, the group with the higher fluorescing values is selected as the population. However, if the Select option is Negative, the group without fluorescence values are chosen as the population.
NumberOfColonies
Default Calculation: If Select is Min or Max, up to 10 colonies are selected, otherwise All colonies are selected.
NumberOfDivisions
The number of partitions to group colonies into. Partitions are ordered by the Unit Operation feature. If the Select option is Min or Max, the lowest or highest partition will be used, respectively.
PopulationName
Default Calculation: By default, each population is labeled as ColonySelection with sequential integers.
Include
Exclude
BlueWhiteScreen
blueWhiteScreenRules
Key/value pairs that define a set of colonies into a population based on their BlueWhiteScreen feature.
Characterization
Color
The color to block with the absorbance filter which is inserted between the light source and the sample. Default to blue to eliminate blue colonies from the image, even when the colonies are newly formed and appear powder blue.
FilterWavelength
The mean wavelength of light blocked by the absorbance filter. Blue colonies, which have a wavelength around 450 nanometers, appear darker.
Select
The method used to group colonies into a population. Colonies are first ordered by their intensities from BlueWhiteScreen images, the average pixel value inside the colony boundary from an image generated using an absorbance filter. If the Select option is Positive or Negative, the ordered colonies are first clustered into two groups based on their Absorbance. Then, if the Select option is Positive, the group with the higher brightness is selected as the population, and in this case white colonies. However, if the Select option is Negative, the group with the lower brightness values are chosen as the population, and in this case blue colonies. Alternatively, if the Select option is Min or Max, the ordered colonies are divided into partitions based on the NumberOfDivisions. From there, the least bright or most bright partition is grouped into a population depending on if the Select option is Min or Max. For example, if the Select option is set to Max and the NumberOfDivisions is 10, the colonies will be divided into 10 partitions sorted from least bright to most bright and the most bright partition will be selected as the population.
NumberOfColonies
Default Calculation: If Select is Min or Max, up to 10 colonies are selected, otherwise All colonies are selected.
NumberOfDivisions
The number of partitions to group colonies into. Partitions are ordered by the Unit Operation feature. If the Select option is Min or Max, the lowest or highest partition will be used, respectively.
PopulationName
Default Calculation: By default, each population is labeled as ColonySelection with sequential integers.
Include
Exclude
AllColonies
MultiFeatured
multiFeaturedRules
Key/value pairs that define a set of colonies into a population by multiple features, including Diameter, Regularity, Isolation, Circularity, BlueWhiteScreen, and Fluorescence.
Characterization
Features
The characteristics selected from Diameter, Regularity, Isolation, Circularity, BlueWhiteScreen, and Fluorescence of the colony by which the population will be isolated. For example, if Features is set to {Isolation, Diameter}, the colonies that are both larger than the median colony and more fluorescing than the median colony will be grouped into a population. More than one feature must be specified, otherwise the individual feature Unit Operation should be used. Colonies that match all features in the MultiFeatured unit operation will be included in the population.
Pattern Description: Diameter, Isolation, Regularity, Circularity, Fluorescence, BlueWhiteScreen, or Absorbance or {Isolation, Diameter}
Select
The method used to group colonies into a population. Colonies are first ordered by their Feature. If the Select option is Min or Max, the ordered colonies are divided into partitions based on the NumberOfDivisions. From there, the smallest or largest partition is grouped into a population depending on if the Select option is Min or Max. For example, if the Select option is set to Max and the NumberOfDivisions is 10, the colonies will be divided into 10 partitions sorted by the Feature and the highest partition will become the selected population. If the Select option is AboveThreshold or BelowThreshold, an absolute cutoff is used to group ordered colonies into a population. The threshold values is set by the Threshold option. For example, if the Select option is BelowThreshold, the Feature is Diameter, and the Threshold is 0.8 Millimeter, all colonies with Diameter less than 0.8 Millimeter will be selected as the population. If the Select option is Positive or Negative, the colonies are first clustered into two groups based on their Fluorescence or BlueWhiteScreen image intensity compared with background. Then, if the Select option is Positive, the group with the higher intensity values is selected as the population. However, if the Select option is Negative, the group without Fluorescence or BlueWhiteScreen signal is chosen as the population.
Default Calculation: For Diameter, Isolation, Regularity, and Circularity features the default is Max unless a Threshold is specified, in which case the default is AboveThreshold. For Fluorescence and BlueWhiteScreen the default is Positive, unless the NumberOfDivisions is specified, in which case the default is Max.
Programmatic Pattern: (Min | Max | AboveThreshold | BelowThreshold | Positive | Negative) | Automatic
NumberOfDivisions
The number of partitions to group colonies into. Partitions are ordered by the Features. If the Select option is Min or Max, the lowest or highest partition will be used, respectively.
Threshold
The min or max possible feature value (e.g., Diameter or Regularity) for a colony to be included in the population. When the Select option is AboveThreshold or BelowThreshold, all colonies with a feature value larger or smaller than the Threshold, respectively, are grouped into a population.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or greater than or equal to 0 millimeters or Null.
Color
The color to block with the absorbance filter which is inserted between the light source and the sample. Default to blue to eliminate blue colonies from the image, even when the colonies are newly formed and appear powder blue.
FilterWavelength
The mean wavelength of light blocked by the absorbance filter. Blue colonies, which have a wavelength around 450 nanometers, appear darker.
Dye
The coloring used to detect colonies that fluoresce at an EmissionWavelength when exposed to an ExcitationWavelength.
Default Calculation: Automatically set to the fluorophore within the excitation/emission range. For VioletFluorescence, set to DAPI. For GreenFluorescence, set to GFP. For OrangeFluorescence, set to Cy3. For RedFluorescence, set to TxRed. For DarkRedFluorescence, set to Cy5.
Pattern Description: DAPI, UltraViolet, GFP, RGBColor[0, 0, 1], Cy3, TxRed, Cy5, or RGBColor[1, 0, 0] or Null.
Programmatic Pattern: ((DAPI | UltraViolet | GFP | Blue | Cy3 | TxRed | Cy5 | Red) | Automatic) | Null
ExcitationWavelength
The wavelength of light that adds energy to colonies and causes them to fluoresce. Pairs with an EmissionWavelength.
Default Calculation: Automatically set to the excitation wavelength specified in the FluorescentExcitationWavelength field of model cell in CellTypes.
Programmatic Pattern: ((377*Nanometer | 457*Nanometer | 531*Nanometer | 628*Nanometer) | Automatic) | Null
EmissionWavelength
The light that the instrument measures to detect colonies fluorescing at a particular wavelength. Pairs with an ExcitationWavelength.
Default Calculation: Automatically set to the emission wavelength specified in the FluorescentEmissionWavelength field of model cell in CellTypes.
Pattern Description: 447 nanometers, 536 nanometers, 593 nanometers, 624 nanometers, or 692 nanometers or Null.
Programmatic Pattern: ((447*Nanometer | 536*Nanometer | 593*Nanometer | 624*Nanometer | 692*Nanometer) | Automatic) | Null
PopulationName
Default Calculation: By default, each population is labeled as ColonySelection with sequential integers.
NumberOfColonies
Default Calculation: If Select is Min or Max, up to 10 colonies are selected, otherwise All colonies are selected.
Include
Exclude
Experiment Options
General
Instrument
The robotic instrument that is used to transfer colonies incubating on solid media to fresh liquid or solid media.
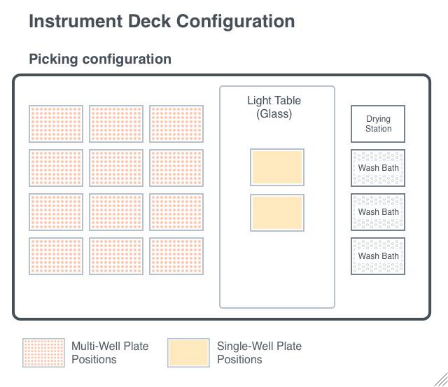
Figure 3.1: Diagram of the deck layout of the ColonyHandler. The ColonyHandler is equipped with colony picking tool. In the picking configuration, the deck features designated positions for multi-well plates, single-well plates inside of light table, and wash baths.
Pattern Description: An object of type or subtype Model[Instrument, ColonyHandler] or Object[Instrument, ColonyHandler]
Programmatic Pattern: ObjectP[{Model[Instrument, ColonyHandler], Object[Instrument, ColonyHandler]}] | Automatic
NumberOfReplicates
The number of times a colony selection should be picked from the input and placed in or on liquid or solid media.
Preparation
Indicates if this unit operation is carried out primarily robotically or manually. Manual unit operations are executed by a laboratory operator and robotic unit operations are executed by a liquid handling work cell.
WorkCell
Characterization
Populations
For each sample, the criteria used to group colonies together into a population to pick. Criteria are based on the ordering of colonies by the desired feature(s): Diameter, Regularity, Circularity, Isolation, Fluorescence, and BlueWhiteScreen. Additionally, CustomCoordinates can be specified, which work in conjunction with the PickCoordinates option to select colonies based on pre-determined locations. For more information see documentation on colony population Unit Operations: Diameter, Isolation, Regularity, Circularity, Fluorescence, BlueWhiteScreen, MultiFeatured, and AllColonies under Experiment Principles section.
Default Calculation: If the Model[Cell] information in the sample object matches one of the fluorescent excitation and emission pairs of the colony picking instrument, Populations is set to Fluorescence. Otherwise, Populations is set to All.
Pattern Description: AllColonies or BlueWhiteScreen or Circularity or Diameter or Fluorescence or Fluorescence, BlueWhiteScreen, Diameter, Isolation, Circularity, Regularity, or All or Isolation or Known Coordinates or MultiFeatured or Regularity.
Programmatic Pattern: ((Fluorescence | BlueWhiteScreen | Diameter | Isolation | Circularity | Regularity | All) | FluorescencePrimitiveP | BlueWhiteScreenPrimitiveP | DiameterPrimitiveP | IsolationPrimitiveP | CircularityPrimitiveP | RegularityPrimitiveP | AllColoniesPrimitiveP | MultiFeaturedPrimitiveP | CustomCoordinates) | Automatic
Selection
MinDiameter
For each sample, the smallest diameter value from which colonies will be included. The diameter is defined as the diameter of a circle with the same area as the colony.
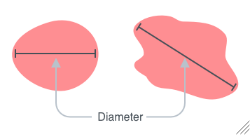
Figure 3.2: Diameter is the distance of the longest line connecting 2 points on the edge of the colony that goes through the centerpoint.
Pattern Description: Greater than or equal to 0.2 millimeters and less than or equal to 10 millimeters or Null.
MaxDiameter
For each sample, the largest diameter value from which colonies will be included. The diameter is defined as the diameter of a circle with the same area as the colony.

Figure 3.3: Diameter is the distance of the longest line connecting 2 points on the edge of the colony that goes through the centerpoint.
Pattern Description: Greater than or equal to 0.2 millimeters and less than or equal to 10 millimeters or Null.
MinColonySeparation
For each sample, the closest distance included colonies can be from each other from which colonies will be included. The separation of a colony is the shortest path between the perimeter of the colony and the perimeter of any other colony.
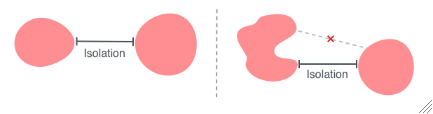
MinRegularityRatio
For each sample, the smallest regularity ratio from which colonies will be included. The regularity ratio is the ratio of the area of the colony to the area of a circle with the colony's perimeter. For example, jagged edged shapes will have a longer perimeter than smoother ones and therefore a smaller regularity ratio.
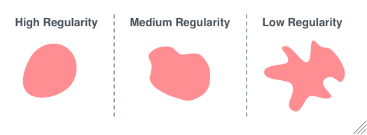
Figure 3.5: Regularity is the ratio of the area of the colony to the area of a circle with the colony's perimeter.
MaxRegularityRatio
For each sample, the largest regularity ratio from which colonies will be included. The regularity ratio is the ratio of the area of the colony to the area of a circle with the colony's perimeter. For example, jagged edged shapes will have a longer perimeter than smoother ones and therefore a smaller regularity ratio.

Figure 3.6: Regularity is the ratio of the area of the colony to the area of a circle with the colony's perimeter.
MinCircularityRatio
For each sample, the smallest circularity ratio from which colonies will be included. The circularity ratio is defined as the ratio of the minor axis to the major axis of the best fit ellipse. For example, a very oblong colony will have a much larger major axis compared to its minor axis and therefore a low circularity ratio.
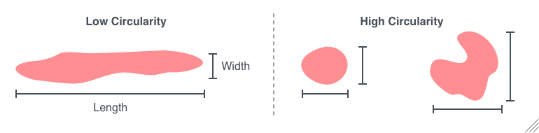
Figure 3.7: Circularity is a measure of the ratio of the minor axis to the major axis of the best fit ellipse.
MaxCircularityRatio
For each sample, the largest circularity ratio from which colonies will be included. The circularity ratio is defined as the ratio of the minor axis to the major axis of the best fit ellipse. For example, a very oblong colony will have a much larger major axis compared to its minor axis and therefore a low circularity ratio.

Figure 3.8: Circularity is a measure of the ratio of the minor axis to the major axis of the best fit ellipse.
Picking
ColonyPickingTool
For each sample, the tool used to stab the source colonies plates from SamplesIn and either deposit any material stuck to the picking head onto the destination plate or into a destination well.
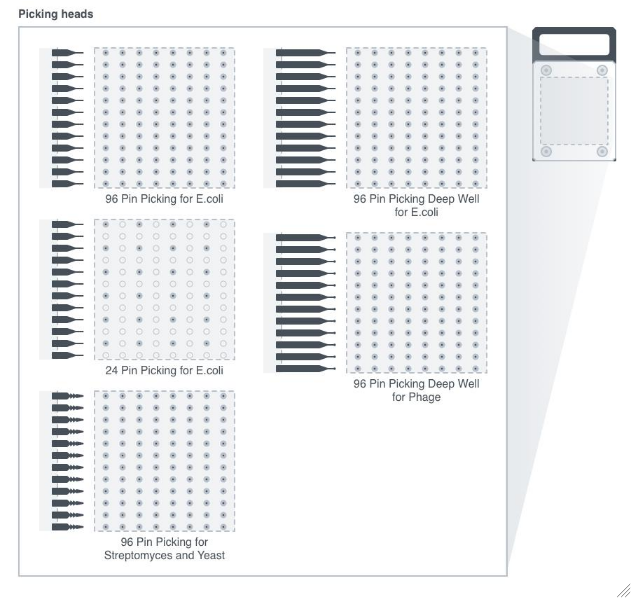
Figure 3.9: Various colony picking tools. In Picking application, the ColonyHandlerHeadCassette containing either 96/24-pin picking head is installed into the robot gantry, and is designed to stab source colonies and deposit any material stuck to the probe onto the destination plate or into a destination well.
Default Calculation: If the DestinationContainer has 24 wells, set to Model[Part,ColonyHandlerHeadCassette, "24-pin picking head for E. coli" ]. If the DestinationContainer has 96 or 384 wells, or is an OmniTray, will use the PreferredColonyHandlerHeadCassette of the first Model[Cell] in the composition of the input sample. If the Composition field is not filled or there are not Model[Cell]'s in the composition, this option is automatically set to Model[Part,ColonyHandlerHeadCassette, "96-pin picking head for E. coli - Deep well"] if the destination is a deep well plate and Model[Part,ColonyHandlerHeadCassette, "96-pin picking head for E. coli"] otherwise.
Pattern Description: An object of type or subtype Model[Part, ColonyHandlerHeadCassette] or Object[Part, ColonyHandlerHeadCassette]
Programmatic Pattern: ObjectP[{Model[Part, ColonyHandlerHeadCassette], Object[Part, ColonyHandlerHeadCassette]}] | Automatic
HeadDiameter
For each sample, the width of the metal probe used to stab the source colonies and deposit any material stuck to the probe onto the destination plate or into a destination well.
HeadLength
For each sample, the length of the metal probe used to stab the source colonies and deposit any material stuck to the probe onto the destination plate or into a destination well.
NumberOfHeads
For each sample, the number of metal probes on the ColonyHandlerHeadCassette used to stab the source colonies and deposit any material stuck to the probe onto the destination plate or into a destination well.
ColonyHandlerHeadCassetteApplication
For each sample, the designed use of the ColonyPickingTool used to stab the source colonies and deposit any material stuck to the probe onto the destination plate or into a destination well..
ColonyPickingDepth
Pattern Description: Greater than or equal to 0 millimeters and less than or equal to 20 millimeters.
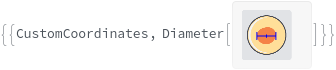
PickCoordinates
For each sample, the coordinates, in Millimeters, from which colonies will be picked from the source plate where {0 Millimeter, 0 Millimeter} is the center of the source well.
Programmatic Pattern: ({{RangeP[-63*Millimeter, 63*Millimeter], RangeP[-43*Millimeter, 43*Millimeter]}..} | Automatic) | Null
Imaging
ImagingStrategies
For each Sample, the end goals for capturing images. ImagingStrategies can be either a single end goal of simple visualization or a list of multiple end goals. The options include BrightField, BlueWhiteScreen, and multiple fluorescence imaging techniques. BrightField imaging is essential for obtaining baseline images and is required if other imaging strategies are employed. BlueWhiteScreen is used to distinguish recombinant colonies with disrupted lacZ (white colonies) from blue colonies using an absorbance filter. Fluorescence imaging allows for the visualization of colonies with fluorescent dyes or proteins.
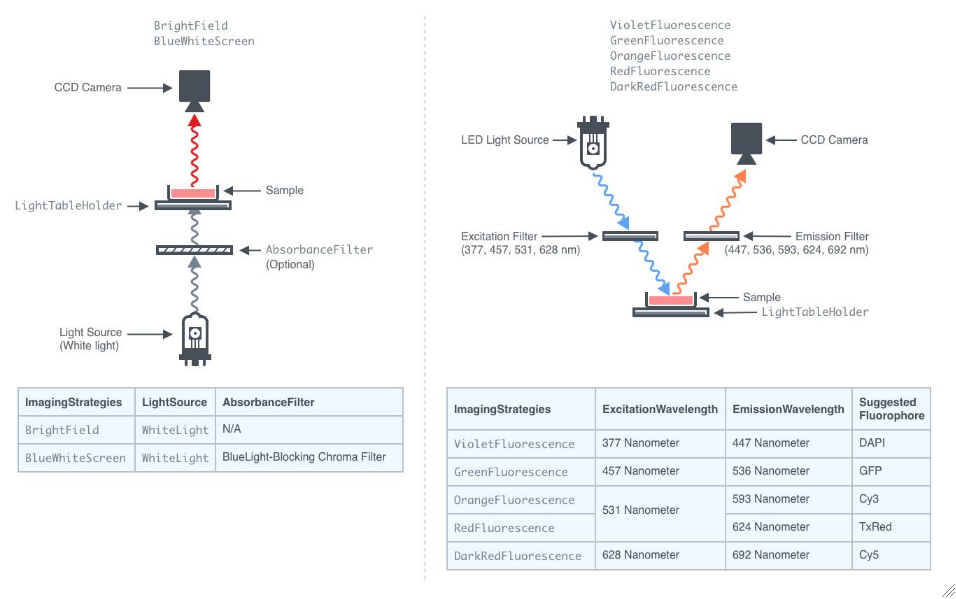
Figure 3.10: Diagram of the imaging module of the ColonyHandler with various imaging strategies. For BlueWhiteScreen, an absorbance filter which blocks blue light between the sample and the light source, eliminating blue colonies from the image, even when they are newly formed and appear powder blue. For VioletFluorescence, GreenFluorescence, OrangeFluorescence, RedFluorescence, or DarkRedFluorescence, an LED-based light source with a specific fluorescence excitation wavelength illuminates the sample, and emission is filtered before reaching the CCD camera. The transition to capture multiple types of images is accomplished by the robotic gantry of the QPix colony handler, which moves seamlessly between imaging strategies and samples.
Default Calculation: Automatically set to include the BlueWhiteScreen along with BrightField if option Populations is set to include BlueWhiteScreen, set to include fluorescence along with BrightField if the Model[Cell] information in the sample matches one of the fluorescent excitation and emission pairs of the imaging instrument. Otherwise, set to BrightField as a BrightField image is always taken.
Programmatic Pattern: (DuplicateFreeListableP[BrightField | BlueWhiteScreen | VioletFluorescence | GreenFluorescence | OrangeFluorescence | RedFluorescence | DarkRedFluorescence] | BrightField) | Automatic
ExposureTimes
For each Sample, and for each imaging strategy, a single length of time to allow the camera to capture an image. An increased exposure time leads to brighter images based on a linear scale. When set as Automatic, optimal exposure time is automatically determined during experiment. This is done by running AnalyzeImageExposure on images taken with suggested initial exposure times. The process adjusts the exposure time for subsequent image acquisitions until the optimal exposure time is found.
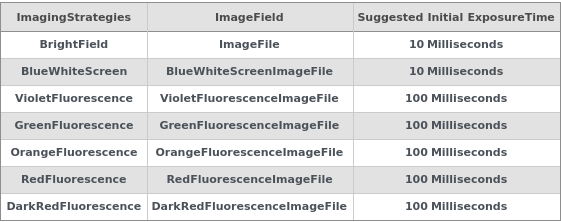
Figure 3.11: This table outlines the suggested exposure times for various imaging strategies and lists the corresponding fields where the raw image data is stored for each imaging strategy.
Programmatic Pattern: ({(RangeP[1*Millisecond, 2000*Millisecond] | Automatic)..} | (RangeP[1*Millisecond, 2000*Millisecond] | Automatic)) | Automatic
Placing
DestinationMediaType
Default Calculation: Automatically set to State that matches DestinationMedia if specified. Otherwise is set to LiquidMedia if there is a PreferredLiquidMedia specified in the Model[Cell] in the input sample. If there is no PreferredLiquidMedia but there is a PreferredSolidMedia, resolves to SolidMedia. If there is neither PreferredLiquidMedia or PreferredSolidMedia in the Model[Cell] in the input sample, defaults to LiquidMedia.
DestinationMedia
Default Calculation: If DestinationMediaType is LiquidMedia, automatically set to the value in the PreferredLiquidMedia field for the first Model[Cell] in the input sample Composition. If DestinationMediaType is SolidMedia, automatically set to the value in the PreferredSolidMedia field for the first Model[Cell] in the input sample.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
DestinationMediaContainer
Default Calculation: Automatically set based on the DestinationMediaType and DestinationMedia options. Will default to Model[Container, Plate, "96-well 2mL Deep Well Plate"] if DestinationMediaType is LiquidMedia and will default to Model[Container, Plate, "Omni Tray Sterile Media Plate"] if DestinationMediaType is SolidMedia.
Programmatic Pattern: (ObjectP[Model[Container]] | (ObjectP[Object[Container]] | _String) | {(ObjectP[Object[Container]] | _String)..}) | Automatic
DestinationFillDirection
For each Sample, indicates if the DestinationMediaContainer is filled with picked colonies in row order, column order, or by custom coordinates. Row/Column completely fills spots in available row/column before moving to the next. If set to CustomCoordinates, ignores MaxDestinationNumberOfColumns and MaxDestinationNumberOfRows to fill at locations specified by DestinationCoordinates. CustomCoordinates is only applicable when DestinationMediaType is SolidMedia.
MaxDestinationNumberOfColumns

Figure 3.12: Picked colonies will be deposited on solid media and spaced out according to this chart.
Default Calculation: Automatically set based on the below table after the number of colonies to pick has been determined.
MaxDestinationNumberOfRows
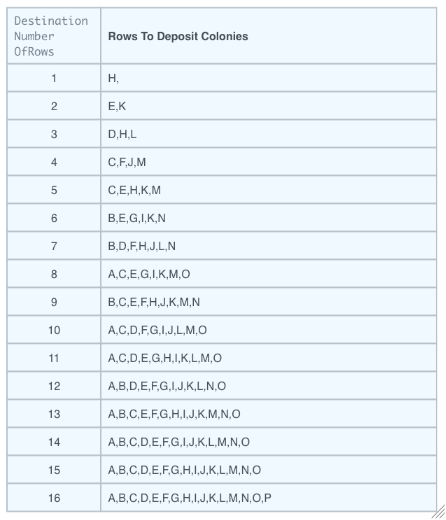
Figure 3.13: Picked colonies will be deposited on solid media and spaced out according to this chart.
Default Calculation: Automatically set based on the below table after the number of colonies to pick has been determined.
DestinationCoordinates
For each sample, the xy coordinates, in Millimeters, to deposit the picked colonies on solid media where {0 Millimeter, 0 Millimeter} is the center of the destination well. DestinationCoordinates is only applicable when Destination is SolidMedia.
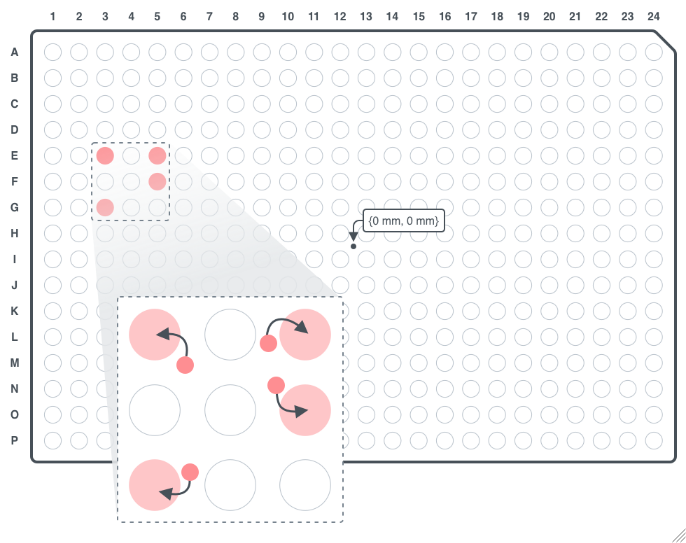
Figure 3.14: The specified coordinates will snap to the center of a 384 well grid on the solid media.
Programmatic Pattern: {{RangeP[-60*Millimeter, 60*Millimeter], RangeP[-40*Millimeter, 40*Millimeter]}..} | Null
MediaVolume
For each sample, the amount of liquid media in which the picked colonies are placed. MediaVolume is only applicable when DestinationMediaType -> LiquidMedia.
Default Calculation: Automatically set to the minimum of $MaxRoboticSingleTransferVolume and either the RecommendedFillVolume of the DestinationMediaContainer, or 40% of the MaxVolume if the field is not populated.
DestinationMix
For each sample, indicates if the picking head is swirled in the destination plate while inoculating the liquid media. DestinationMix is only applicable when Destination -> LiquidMedia.
Default Calculation: If Destination is LiquidMedia or if DestinationNumberOfMixes is specified, automatically resolves to True.
DestinationNumberOfMixes
For each sample, the number of times the picking pin will be swirled in the liquid media during inoculation. DestinationNumberOfMixes is only applicable when Destination -> LiquidMedia.
Pattern Description: Greater than or equal to 0 and less than or equal to 50 in increments of 1 or Null.
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be disposed.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal.
SamplesOutStorageCondition
The non-default conditions under which any new samples generated by this experiment should be stored after the protocol is completed. If left unset, the new samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Sanitization
PrimaryWash
For each sample, indicates if the PrimaryWash stage should be turned on during the sanitization process.
PrimaryWashSolution
For each sample, the first wash solution that is used during the sanitization process prior to each round of picking.
Default Calculation: Automatically set to Model[Sample,StockSolution,"70% Ethanol"], if PrimaryWash is True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
NumberOfPrimaryWashes
For each sample, the number of times the ColonyHandlerHeadCassette moves in a circular motion in the PrimaryWashSolution to clean any material off the head.
PrimaryDryTime
For each sample, the length of time the ColonyHandlerHeadCassette is dried over the halogen fan after the cassette is washed in PrimaryWashSolution.
SecondaryWash
For each sample, indicates if the SecondaryWash stage should be turned on during the sanitization process.
SecondaryWashSolution
For each sample, the second wash solution that can be used during the sanitization process prior to each round of picking.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
NumberOfSecondaryWashes
For each sample, the number of times the ColonyHandlerHeadCassette moves in a circular motion in the SecondaryWashSolution to clean any material off the head.
SecondaryDryTime
For each sample, the length of time the ColonyHandlerHeadCassette is dried over the halogen fan after the cassette is washed in SecondaryWashSolution.
TertiaryWash
For each sample, indicates if the TertiaryWash stage should be turned on during the sanitization process.
TertiaryWashSolution
For each sample, the third wash solution that can be used during the sanitization process prior to each round of picking.
Default Calculation: Automatically set to Model[Sample, StockSolution, "10% Bleach"] if TertiaryWash is True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
NumberOfTertiaryWashes
For each sample, the number of times the ColonyHandlerHeadCassette moves in a circular motion in the TertiaryWashSolution to clean any material off the head.
TertiaryDryTime
For each sample, the length of time to dry the ColonyHandlerHeadCassette over the halogen fan after the cassette is washed in TertiaryWashSolution.
QuaternaryWash
For each sample, indicates if the QuaternaryWash stage should be turned on during the sanitization process.
QuaternaryWashSolution
For each sample, the fourth wash solution that can be used during the process prior to each round of picking.
Default Calculation: Automatically set to Model[Sample,StockSolution,"70% Ethanol"] if QuaternaryWash is True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
NumberOfQuaternaryWashes
For each sample, the number of times the ColonyHandlerHeadCassette moves in a circular motion in the QuaternaryWashSolution to clean any material off the head
QuaternaryDryTime
Protocol Options
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Indicates if any samples that are modified in the course of the experiment should be freshly imaged after running the experiment. Please note that public samples are imaged regardless of the value of this option.
Organizational Information
Name
Example Calls
Picking


Pick colonies after they have been classified by multiple features. In this case the large fluorescing colonies would be picked:

Inoculate Media
Have the picked colonies placed into LiquidMedia and then mixed by pipette, so they can be further grown:

Have the picked colonies placed onto SolidMedia by specifying a custom placement pattern, so they can be further grown. {0 Millimeter, 0 Millimeter is the center of the destination well}:
Warnings and Errors
Messages (46)
ColonyHandlerHeadCassetteApplicationMismatch (1)
DestinationFillDirectionMismatch (1)
DestinationMediaTypeMismatch (1)
DestinationMixMismatch (1)
HeadDiameterMismatch (1)
HeadLengthMismatch (1)
ImagingOptionMismatch (2)
IncompatibleMaterials (2)
IndexMatchingPrimitive (1)
InstrumentPrecision (6)
If a ColonyPickingDepth with a greater precision than 0.01 Millimeter is given, it is rounded:


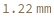
If an ExposureTime with a greater precision than 1 Millisecond is given, it is rounded:


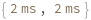
If a PrimaryDryTime with a greater precision than 1 Second is given, it is rounded:


If a QuaternaryDryTime with a greater precision than 1 Second is given, it is rounded:


If a SecondaryDryTime with a greater precision than 1 Second is given, it is rounded:


If a TertiaryDryTime with a greater precision than 1 Second is given, it is rounded:


InvalidColonyPickingDepths (1)
InvalidDestinationMediaContainer (1)
InvalidDestinationMediaState (1)
InvalidMixOption (1)
MissingDestinationCoordinates (1)
MissingImagingStrategies (1)
MultiplePopulationMethods (1)
NoAutomaticWavelength (1)
NoAvailablePickingTool (1)
NotPreferredColonyHandlerHead (1)
NumberOfHeadsMismatch (1)
OutOfOrderWashStages (1)
OverlappingPopulations (1)
PickCoordinatesMissing (1)
PickingToolIncompatibleWithDestinationMediaContainer (1)
PrimaryWashMismatch (1)
QPixWashSolutionInsufficientVolume (1)
QuaternaryWashMismatch (1)
RepeatedPopulationNames (1)
SecondaryWashMismatch (1)
SingleAutomaticWavelength (1)
TertiaryWashMismatch (1)
TooManyDestinationCoordinates (1)
TooManyDestinationMediaContainers (1)
TooManyInputContainers (1)
Last modified on Sat 2 Nov 2024 18:51:23