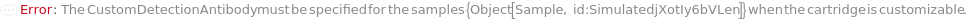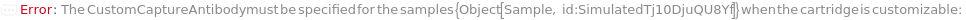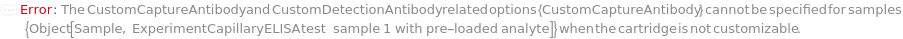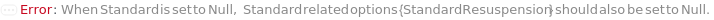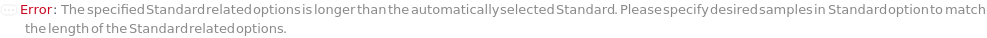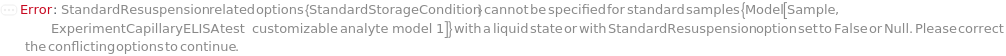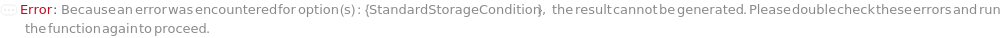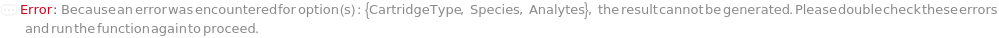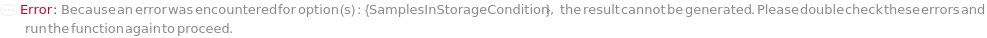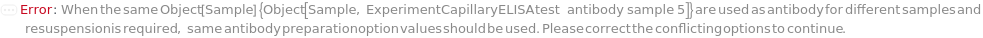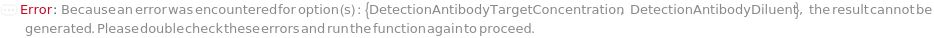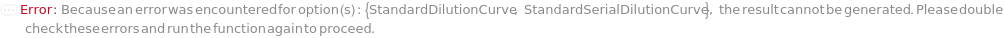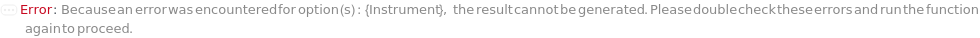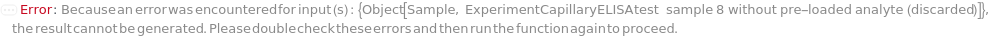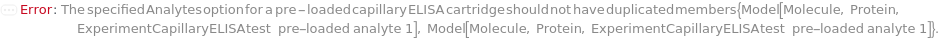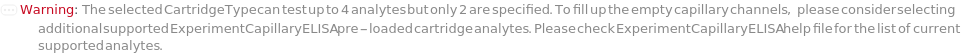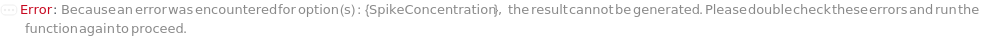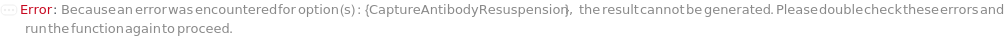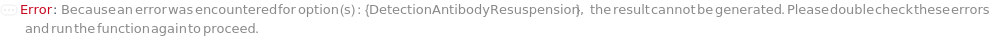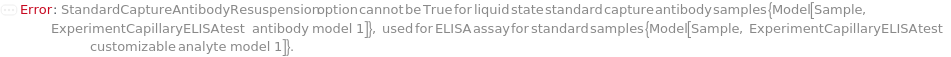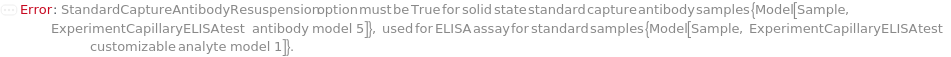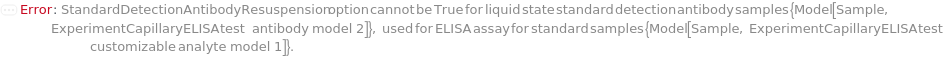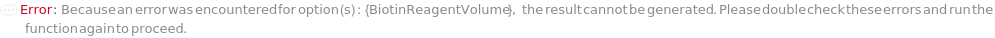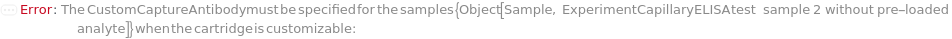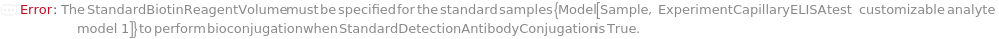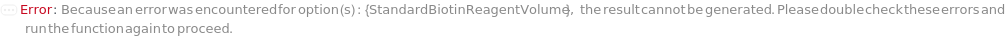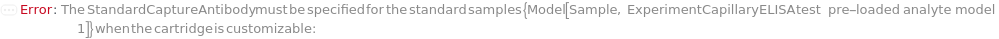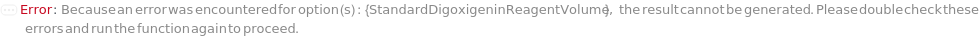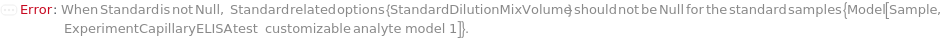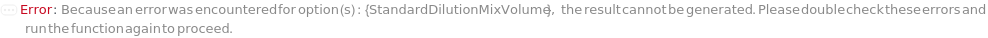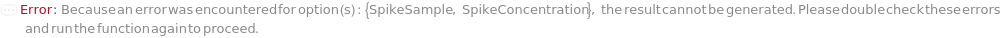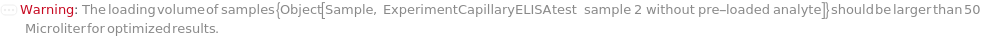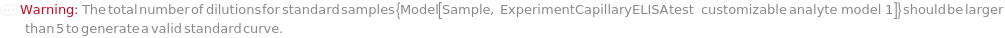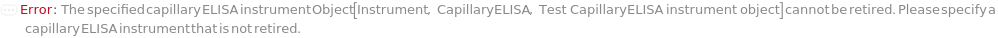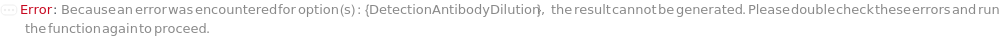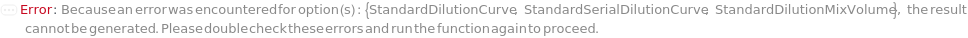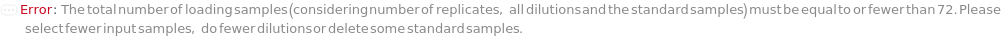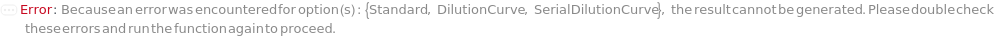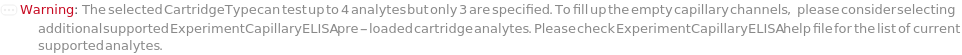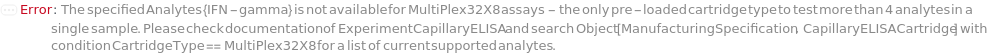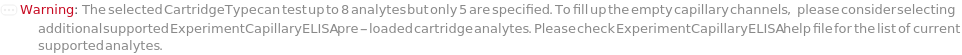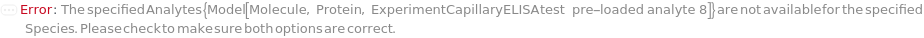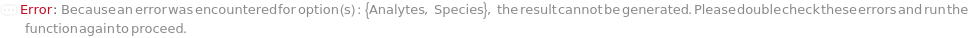General
Instrument
The detection and quantification capillary ELISA device on which the protocol is to be run. The instrument accepts the cartridge loaded with samples and buffers, loads the samples into the capillaries, performs the ELISA experiment and detects the signals to quantify certain analytes.
Default Value: Model[Instrument, CapillaryELISA, id:XnlV5jKM7zPM]
Pattern Description: An object of type or subtype Model[Instrument, CapillaryELISA] or Object[Instrument, CapillaryELISA]
Programmatic Pattern: ObjectP[{Model[Instrument, CapillaryELISA], Object[Instrument, CapillaryELISA]}]
CartridgeType
The type of capillary ELISA cartridge (SinglePlex72X1, MultiAnalyte32X4, MultiAnalyte16X4, MultiPlex32X8 or Customizable) that is used in the capillary ELISA experiment. For pre-loaded cartridge types, different cartridge types provide different numbers of capacity and throughput (Figure 3.1 in ExperimentCapillaryELISA help file).
Default Calculation: CartridgeType is automatically selected based on the total number of samples (with standard samples and dilutions considered) and the Cartridge option. When the total number of samples is below or equal to 48 and Cartridge option is not Null, CartridgeType is set to Customizable. Otherwise it is set to SinglePlex72X1.
Pattern Description: SinglePlex72X1, MultiAnalyte32X4, MultiAnalyte16X4, MultiPlex32X8, or Customizable.
Programmatic Pattern: ELISACartridgeTypeP | Automatic
Cartridge
The single-use capillary ELISA cartridge plate that is used in the instrument to provide the entire workflow of a sandwich ELISA assay and quantify the analytes (such as peptides, proteins, antibodies and hormones) in the samples. A customizable capillary ELISA cartridge Model[Container,Plate,Irregular,CapillaryELISA,"Human 48-Digoxigenin Cartridge"] uses an anti-digoxigenin antibody coating in GNR to serve as the foundation. Digoxigenin-modified capture antibody sample and biotinylated detection antibody sample are prepared prior to the experiment and loaded into the Cartridge for sandwich ELISA experiment of up to 48 samples. For pre-loaded cartridge types (all types except Customizable), the cartridge is loaded with capture antibody and detection antibody reagents, meaning that Cartridge option also determines the ELISA assay to be run and the analytes to be detected in the experiment. For the current list of available capillary ELISA cartridges, please perform Search[Model[Container,Plate,Irregular,CapillaryELISA]] with the desired CartridgeType, AnalyteMolecules or other conditions. If a new model of pre-loaded cartridge is required for the experiment, please set Cartridge option to Null and input more information in CartridgeType, Species and Analytes options. The new model is created and the cartridge is fulfilled accordingly. For a complete list of available analytes on pre-loaded cartridges and their ordering information, please refer to Object[Report,Literature,"Simple Plex Assay Menu"] and Object[ManufacturingSpecification,CapillaryELISACartridge] of different analytes for more details.
Default Calculation: Cartridge option is automatically set to an in-stock pre-loaded cartridge object that matches the analytes presented in the samples. If no in-stock cartridge is available for the specified options and the total number of samples is below 48, Cartridge option is automatically resolved to the customizable capillary ELISA cartridge to eliminate material waiting time. Otherwise, a pre-loaded capillary ELISA cartridge model is selected or created, with an estimated lead time of 14 days after protocol confirmation.
Pattern Description: An object of type or subtype Model[Container, Plate, Irregular, CapillaryELISA] or Object[Container, Plate, Irregular, CapillaryELISA] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Irregular, CapillaryELISA], Object[Container, Plate, Irregular, CapillaryELISA]}] | Automatic) | Null
Species
The organism (human, mouse or rat) that the samples (containing analytes of interest) are derived from. This option is used to determine the available analytes for pre-loaded capillary ELISA cartridges.
Pattern Description: Human, Mouse, Rat, or Other.
Programmatic Pattern: ELISASpeciesP
Analytes
The target(s) (e.g., peptides, proteins, antibodies, hormones) detected and quantified in each experiment sample. For customizable cartridge, only 1 analyte can be detected per sample. A pre-loaded cartridge is loaded with complete assay materials (capture antibody, detection antibody and fluorescent labelling reagent) for the selected analytes and all the samples are subject to the same analytes. For a complete list of available analytes of pre-loaded cartridges, please refer to Object[Report,Literature,"Simple Plex Assay Menu"]. To create a capillary ELISA cartridge with combination of multiple targets, please refer to Object[ManufacturingSpecification,CapillaryELISACartridge] of different analytes for more details about compatibility rules and ordering information. For SinglePlex72X1 cartridge type, only 1 analyte can be selected. For MultiAnalyte32X4 and MultiAnalyte16X4 cartridge types, up to 4 distinct analytes can be selected. For MultiPlex32X8 cartridge type, up to 8 analytes can be selected in Pro-Inflammation and Oncology panel only.
Default Calculation: If a customizable cartridge is used, Analytes option is automatically set based on the presented analyte in the Analytes field or Composition field of each input sample. If Cartridge option is set to an existing model or object of pre-loaded capillary ELISA cartridge plate, Analytes option is automatically set to the Analytes of Cartridge option. If Cartridge option is set to Null, a new pre-loaded model of capillary ELISA cartridge is requested. In this case, Analytes option is set based on the selected CartridgeType, Species and presented analytes of the input sample objects (in Analytes field or Composition field).
Pattern Description: Multiplex or Single Analyte or Null.
Programmatic Pattern: (((ObjectP[{Model[Molecule]}] | CapillaryELISAAnalyteP) | ({ObjectP[{Model[Molecule]}]..} | {CapillaryELISAAnalyteP..})) | Automatic) | Null
Index Matches to: experiment samples
NumberOfReplicates
Number of times each of the input samples should be analyzed using identical experimental parameters.
Pattern Description: Greater than or equal to 2 in increments of 1 or Null.
Programmatic Pattern: GreaterEqualP[2, 1] | Null
Sample Preparation
SampleVolume
The initial sample amount aliquoted from the source of the input sample. If a SpikeSample is specified, this it the volume of the input sample to mix with SpikeVolume of SpikeSample before further dilution is performed.
Default Calculation: SampleVolume is automatically set to 30 Microliter when SpikeSample is not Null.
Pattern Description: Greater than 1 microliter or Null.
Programmatic Pattern: (GreaterP[1*Microliter] | Automatic) | Null
Index Matches to: experiment samples
SpikeSample
For each experiment sample, the sample with known concentration(s) of analyte(s) mixed with the input sample to increase the analytical response of the unknown sample, for evaluation of sample matrices in a new ELISA assay.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or Null.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | Null
Index Matches to: experiment samples
SpikeVolume
The volume of the SpikeSample mixed with the SampleVolume of the input sample before further dilution is performed.
Default Calculation: SpikeVolume is automatically set to SampleVolume when SpikeSample is not Null.
Pattern Description: Greater than 1 microliter or Null.
Programmatic Pattern: (GreaterP[1*Microliter] | Automatic) | Null
Index Matches to: experiment samples
DilutionCurve
The collection of dilutions performed on each input sample (plus SpikeSample, if applicable). Dilution is required on all the loading samples for the best performance of microfluidics and the ELISA experiment is performed on all the diluted samples. For FixedDilutionVolume dilutions, the SampleAmount is the volume of the sample that is mixed with the DiluentVolume of the Diluent. For FixedDilutionFactor dilutions, the AssayVolume is the volume of the sample that is created after being diluted by the specified DilutionFactor. For a pre-loaded capillary ELISA cartridge, please refer to Object[ManufacturingSpecification,CapillaryELISACartridge] of the analytes for their recommended minimum dilution factors. For a customizable cartridge, a minimum dilution factor of 0.5 is suggested.
Default Calculation: If SerialDilutionCurve is not uploaded, DilutionCurve option is automatically set to 1 dilution with DilutionFactor of the minimum required dilution factor to generate a AssayVolume of 60 Microliter.
Pattern Description: Fixed Dilution Factor or Fixed Dilution Volume or Null.
Programmatic Pattern: (({{GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter]}..} | {{GreaterEqualP[30*Microliter], RangeP[0, 1]}..}) | Automatic) | Null
Index Matches to: experiment samples
SerialDilutionCurve
The collection of stepwise dilutions performed on each input sample (plus SpikeSample, if applicable). Dilution is required on all the loading samples for the best performance of microfluidics and the ELISA assay is performed on all the diluted samples. To use non-diluted input sample, the first dilution can be set to a dilution factor of 1. For SerialDilutionVolumes, the TransferVolume is taken out of the sample and added to a second well with the DiluentVolume of the Diluent. After thorough mixing, the TransferVolume is taken out of that well and added to a third well. This is repeated to make NumberofDilutions diluted samples. For SerialDilutionFactors, the sample is diluted by the dilution factor at each transfer step. For a pre-loaded capillary ELISA cartridge, please refer to Object[ManufacturingSpecification,CapillaryELISACartridge] of the analytes for their recommended minimum dilution factors. For a customizable cartridge, a minimum dilution factor of 0.5 is suggested.
Default Calculation: If DilutionCurve option is Null, SerialDilutionCurve option is automatically set to 1 NumberOfDilutions with ConstantDilutionFactor of the recommended minimum dilution factor to generate a AssayVolume of 60 Microliter.
Pattern Description: Serial Dilution Factor or Serial Dilution Volumes or Null.
Programmatic Pattern: (({GreaterP[0*Microliter], GreaterEqualP[30*Microliter], GreaterP[1, 1]} | {GreaterEqualP[30*Microliter], {RangeP[0, 1], GreaterP[1, 1]} | {RangeP[0, 1]..}}) | Automatic) | Null
Index Matches to: experiment samples
Diluent
For each experiment sample, the diluent buffer used to mix with the input sample (plus SpikeSample, if applicable) to reduce its concentration in order to make DilutionCurve or SerialDilutionCurve. Dilution with the recommended diluent is required on all the loading samples for the best performance of microfluidics.
Default Calculation: If a pre-loaded cartridge is used for the experiment, Diluent option is automatically set to the recommended diluent provided by the assay developer for the specified Analytes or Cartridge. For a customizable cartridge, Diluent option is set to Model[Sample,"Simple Plex Sample Diluent 13"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic
Index Matches to: experiment samples
DilutionMixVolume
The volume that is pipetted up and down in the diluted input sample to mix each sample thoroughly with Diluent to make DilutionCurve or SerialDilutionCurve.
Default Calculation: DilutionMixVolume option is automatically set to half of the minimum prepared volume of the diluted sample
Pattern Description: Greater than or equal to 0 microliters.
Programmatic Pattern: GreaterEqualP[0*Microliter] | Automatic
Index Matches to: experiment samples
DilutionNumberOfMixes
The number of pipette up and down cycles that is used to mix each input sample thoroughly with Diluent to make DilutionCurve or SerialDilutionCurve.
Pattern Description: Greater than or equal to 0 and less than or equal to 20 in increments of 1.
Programmatic Pattern: RangeP[0, 20, 1]
Index Matches to: experiment samples
DilutionMixRate
The speed at which the DilutionMixVolume is pipetted up and down in the diluted input sample to mix each sample thoroughly with Diluent to make DilutionCurve or SerialDilutionCurve.
Default Value: 100 microliters per second
Pattern Description: Greater than or equal to 0.4 microliters per second and less than or equal to 250 microliters per second.
Programmatic Pattern: RangeP[0.4*(Microliter/Second), 250*(Microliter/Second)]
Index Matches to: experiment samples
Post Experiment
SpikeSampleStorageCondition
For each SpikeSample, the non-default conditions under which it should be stored after the protocol is completed. If left unset, the SpikeSample will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: (SampleStorageTypeP | Disposal) | Null
Index Matches to: experiment samples
CaptureAntibodyStorageCondition
Indicates the condition under which the unused portion of liquid state CustomCaptureAntibody sample inside its original container is stored after the protocol is completed.
Default Calculation: CaptureAntibodyStorageCondition option is automatically set to Freezer if CaptureAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyStorageCondition
Indicates the condition under which the unused portion of liquid state CustomDetectionAntibody sample inside its original container is stored after the protocol is completed.
Default Calculation: DetectionAntibodyStorageCondition option is automatically set to Freezer if DetectionAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
StandardStorageCondition
Indicates the condition under which the unused portion of liquid state Standard inside its original container is stored after the protocol is completed.
Default Calculation: StandardStorageCondition option is automatically set to Freezer if StandardResuspension is True.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyStorageCondition
Indicates the condition under which the unused portion of liquid state StandardCaptureAntibody sample inside its original container is stored after the protocol is completed.
Default Calculation: StandardCaptureAntibodyStorageCondition option is automatically set to Freezer if StandardCaptureAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyStorageCondition
Indicates the condition under which the unused portion of liquid state StandardDetectionAntibody sample inside its original container is stored after the protocol is completed.
Default Calculation: StandardDetectionAntibodyStorageCondition option is automatically set to Freezer if StandardDetectionAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: Standard
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: experiment samples
Capture Antibody Preparation
CustomCaptureAntibody
For each experiment sample, the capture antibody sample used in the sandwich ELISA experiment to bind with the present analytes through specific antigen-antibody interaction. The capture antibody must be labeled with digoxigenin, purified and diluted to preferably 3.5 Microgram/Milliliter before loading into the capillary ELISA cartridge. During the experiment, digoxigenin-labeled capture antibody sample first flows in the fluidic circuit to allow its binding to the immobilized anti-digoxigenin monoclonal antibody in the capillaries. Then the diluted input samples are applied to allow the binding to capture antibody for immobilization. The provided CustomCaptureAntibody can be resuspended into solution, bioconjugated with digoxigenin and diluted as desired using the related options. Please note that if the same capture antibody preparation options are specified for multiple samples (standard samples and/or input samples), the preparation of digoxigenin-labeled capture antibody is performed together. Please consider using ExperimentSamplePreparation for more customized capture antibody preparation.
Default Calculation: If a customizable cartridge is used, CustomCaptureAntibody is automatically set based on the Analytes option. The Antibodies or SecondaryAntibodies field in Model[Molecule] of Analytes is inspected. If available, the DefaultSampleModel of a digoxigenin-labeled antibody is automatically selected as CustomCaptureAntibody. If no digoxigenin-labeled antibody is available, the DefaultSampleModel of an antibody with different epitopes with CustomDetectionAntibody (if uploaded) is automatically selected as CustomCaptureAntibody. If neither CustomCaptureAntibody or CustomDetectionAntibody has been specified and two antibodies with different epitopes are available for the analyte, the DefaultSampleModel of a polyclonal antibody is preferably selected for CustomCaptureAntibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyResuspension
Indicates if the CustomCaptureAntibody should be reconstituted with CaptureAntibodyResuspensionDiluent. If set to True, the original container of the CustomCaptureAntibody sample is centrifuged at 3000 RPM for 1 Minute to eliminate sample loss upon cap opening. Then the CaptureAntibodyResuspensionDiluent is delivered into the CustomCaptureAntibody's container and thorough mixing is achieved by inversion for 30 times.
Default Calculation: CaptureAntibodyResuspension is automatically set to True for a solid state CustomCaptureAntibody sample in a Customizable capillary ELISA cartridge.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyResuspensionConcentration
The target final concentration of the resuspended CustomCaptureAntibody sample.
Default Calculation: CaptureAntibodyResuspensionConcentration option is automatically set to 1 Milligram/Milliliter if CaptureAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyResuspensionDiluent
The resuspension buffer mixed with the solid state CustomCaptureAntibody sample to dissolve the sample into solution.
Default Calculation: CaptureAntibodyResuspensionDiluent option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if CaptureAntibodyResuspension is True and a customizable cartridge is used. The amount of the solid state CustomCaptureAntibody sample is used to calculate the amount of CaptureAntibodyResuspensionDiluent needed.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyConjugation
Indicates if bioconjugation reaction between digoxigenin NHS-ester and primary amines of the CustomCaptureAntibody is required to be performed to prepare digoxigenin-labeled capture antibody sample for capillary ELISA experiment. If set to True, the conjugation mixture is vortexed at 1900 RPM for 1 Minute to allow thorough mixing and then incubated on a heat block at CaptureAntibodyConjugationTemperature for CaptureAntibodyConjugationTime to allow complete reaction.
Default Calculation: CaptureAntibodyConjugation is automatically set to True if the antibody composition of the CustomCaptureAntibody in the Customizable capillary ELISA cartridge does not show Model[Molecule,Protein,Antibody,"Anti-Digoxigenin Antibody"] as its SecondaryAntibodies.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyVolume
The volume of the unconjugated CustomCaptureAntibody sample used to react with DigoxigeninReagent to prepare digoxigenin-labeled capture antibody sample through bioconjugation synthesis.
Default Calculation: CaptureAntibodyVolume option is automatically set to 100 Microliter if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
DigoxigeninReagent
The digoxigenin NHS-ester reagent used to react with the primary amines of the unconjugated CustomCaptureAntibody sample to prepare digoxigenin-labeled capture antibody sample through bioconjugation synthesis.
Default Calculation: DigoxigeninReagent option is automatically set to Model[Sample,StockSolution,"Digoxigenin-NHS, 0.67 mg/mL in DMF"] if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
DigoxigeninReagentVolume
The volume of DigoxigeninReagent used to react with CaptureAntibodyVolume of the unconjugated CustomCaptureAntibody sample to prepare digoxigenin-labeled capture antibody sample through bioconjugation synthesis.
Default Calculation: When a customizable cartridge is used, DigoxigeninReagentVolume option is automatically set using the concentration of DigoxigeninReagent and the antibody concentration to achieve a 5:1 molar ratio between DigoxigeninReagent and the unconjugated CustomCaptureAntibody.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyConjugationBuffer
The buffer solution in which the reaction between the unconjugated CustomCaptureAntibody sample and DigoxigeninReagent happens to prepare digoxigenin-labeled capture antibody sample through bioconjugation synthesis.
Default Calculation: CaptureAntibodyConjugationBuffer option is automatically set to Model[Sample,StockSolution,"Sodium bicarbonate working stock 75 mg/mL"] if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyConjugationBufferVolume
The volume of CaptureAntibodyConjugationBuffer used to mix with the unconjugated CustomCaptureAntibody sample and DigoxigeninReagent to provide a buffered environment for the bioconjugation reaction.
Default Calculation: When a customizable cartridge is used, CaptureAntibodyConjugationBufferVolume option is automatically set to 1/9 of total volume of CaptureAntibodyVolume and DigoxigeninReagentVolume.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyConjugationContainer
The container in which the unconjugated CustomCaptureAntibody sample, DigoxigeninReagent and CaptureAntibodyConjugationBuffer react to allow bioconjugation synthesis.
Default Calculation: CaptureAntibodyConjugationContainer option is automatically selected base on the total conjugation reaction volume if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyConjugationTime
The amount of reaction time that the CustomCaptureAntibody and DigoxigeninReagent are allowed to react before purification happens.
Default Calculation: CaptureAntibodyConjugationTime option is automatically set to 1 Hour if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyConjugationTemperature
The temperature at which the reaction between the unconjugated CustomCaptureAntibody and DigoxigeninReagent is conducted.
Default Calculation: CaptureAntibodyConjugationTemperature option is automatically set to Ambient if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[-20*Celsius, 150*Celsius]) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyPurificationColumn
The desalting spin column used to adsorb the molecules that are smaller than its molecular weight cut-off (MWCO) on its resin bed to remove the free digoxigenin NHS-ester molecules in order to purify the synthesis product of the CustomCaptureAntibody bioconjugation reaction. The spin column is operated following the manufacturer's instruction (Figure 3.4 in ExperimentCapillaryELISA help file).
Default Calculation: CaptureAntibodyPurificationColumn is automatically set to a 40K molecular weight cut-off Zeba desalting spin column depending on the total volume of the synthesis reaction, if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container, Vessel, Filter] or Object[Container, Vessel, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Vessel, Filter], Object[Container, Vessel, Filter]}] | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyColumnWashBuffer
The buffer solution loaded into the CaptureAntibodyPurificationColumn after its storage buffer is removed and before the sample is loaded. The CaptureAntibodyColumnWashBuffer flushed through CaptureAntibodyPurificationColumn to remove any residues from the spin column storage solution. The washing process is repeated 3 times to equilibrate the resin bed before sample loading.
Default Calculation: CaptureAntibodyColumnWashBuffer option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyConjugationStorageCondition
Indicates the condition under which the unused portion of the synthesized and purified digoxigenin-labeled capture antibody sample is stored after the protocol is completed.
Default Calculation: CaptureAntibodyConjugationStorageCondition option is automatically set to Refrigerator if CaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyDilution
Indicates if digoxigenin-labeled capture antibody dilution is required before loading into capillary ELISA cartridge.
Default Calculation: When a Customizable capillary ELISA cartridge is used, CaptureAntibodyDilution is automatically set to True if CaptureAntibodyResuspension and/or CaptureAntibodyConjugation is True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyTargetConcentration
The desired concentration of digoxigenin-labeled capture antibody after dilution. The initial mass concentration of digoxigenin-labeled capture antibody sample is acquired from the composition of the liquid state CustomCaptureAntibody sample or the CaptureAntibodyResuspension of the solid state CustomCaptureAntibody sample. A 50% concentration loss is factored in if conjugation is performed in the experiment.
Default Calculation: CaptureAntibodyTargetConcentration is automatically set to 3.5 Microgram/Milliliter if CaptureAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: experiment samples
CaptureAntibodyDiluent
The buffer solution used to mix with the concentrated digoxigenin-labeled capture antibody sample to achieve the desired CaptureAntibodyTargetConcentration for cartridge loading.
Default Calculation: CaptureAntibodyDiluent option is automatically set to Model[Sample, "Simple Plex Reagent Diluent"] if CaptureAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
Detection Antibody Preparation
CustomDetectionAntibody
The detection antibody sample used in the sandwich ELISA experiment. After immobilization of the analytes in the microfluidic channels, the detection antibodies bind to analyte epitopes distinct from the capture antibodies through antigen-antibody interaction. Then the streptavidin-conjugated fluorescent dyes can be attached through biotin-streptavidin interaction for data acquiring. The detection antibody must be modified with biotin, purified and diluted to preferably 3.5 Microgram/Milliliter before loading into the capillary ELISA cartridge. The provided CustomDetectionAntibody can be resuspended into solution, bioconjugated with biotin and diluted as desired using the related options. Please note that if the same detection antibody preparation options are specified for multiple samples (standard samples and/or input samples), the preparation of biotinylated detection antibody is performed together. Please consider using ExperimentSamplePreparation for more customized detection antibody preparation.
Default Calculation: If a customizable cartridge is used, CustomDetectionAntibody is automatically set based on the Analytes option. The Antibodies or SecondaryAntibodies field in Model[Molecule] of Analytes is inspected. If available, the DefaultSampleModel of a biotinylated antibody is automatically selected as CustomDetectionAntibody. If no biotinylated antibody is available, the DefaultSampleModel of an antibody with different epitopes with CustomCaptureAntibody (if specified) is automatically selected as CustomDetectionAntibody. If neither CustomCaptureAntibody or CustomDetectionAntibody has been specified and two antibodies with different epitopes are available for the analyte, the DefaultSampleModel of a monoclonal antibody is preferably selected for CustomDetectionAntibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyResuspension
Indicates if the CustomDetectionAntibody should be reconstituted with DetectionAntibodyResuspensionDiluent. If set to True, the original container of the CustomDetectionAntibody sample is centrifuged at 3000 RPM for 1 Minute to eliminate sample loss upon cap opening. Then the DetectionAntibodyResuspensionDiluent is delivered into the CustomDetectionAntibody's container and thorough mixing is achieved by inversion for 30 times.
Default Calculation: DetectionAntibodyResuspension is automatically set to True for a solid state CustomDetectionAntibody sample in a Customizable capillary ELISA cartridge.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyResuspensionConcentration
The target final concentration of the resuspended CustomDetectionAntibody sample.
Default Calculation: DetectionAntibodyResuspensionConcentration option is automatically set to 1 Milligram/Milliliter if DetectionAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyResuspensionDiluent
The resuspension buffer mixed with the solid state CustomDetectionAntibody sample to dissolve the sample into solution.
Default Calculation: DetectionAntibodyResuspensionDiluent option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if DetectionAntibodyResuspension is True and a customizable cartridge is used. The amount of the solid state CustomDetectionAntibody sample is used to calculate the amount of DetectionAntibodyResuspensionDiluent needed.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyConjugation
Indicates if bioconjugation reaction between Biotin-XX-SE (6-((6-((Biotinoyl)Amino)Hexanoyl)Amino)Hexanoic Acid, Succinimidyl Ester) and primary amines of the CustomDetectionAntibody is required to be performed to prepare biotinylated detection antibody sample for capillary ELISA experiment. If set to True, the conjugation mixture is vortexed at 1900 RPM for 1 Minute to allow thorough mixing and then incubated on a heat block at DetectionAntibodyConjugationTemperature for DetectionAntibodyConjugationTime to allow complete reaction.
Default Calculation: DetectionAntibodyConjugation is automatically set to True if the antibody composition of the CustomDetectionAntibody in the Customizable capillary ELISA cartridge does not show Model[Molecule,Protein,"Streptavidin"] as its Targets.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyVolume
The volume of the unconjugated CustomDetectionAntibody sample used to react with BiotinReagent to prepare biotinylated detection antibody sample through bioconjugation synthesis.
Default Calculation: DetectionAntibodyVolume option is automatically set to 100 Microliter if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
BiotinReagent
The Biotin-XX-SE (6-((6-((Biotinoyl)Amino)Hexanoyl)Amino)Hexanoic Acid, Succinimidyl Ester) or other biotinylation reagent used to mix with the unconjugated CustomDetectionAntibody sample to prepare biotinylated detection antibody sample through bioconjugation synthesis.
Default Calculation: BiotinReagent option is automatically set to Model[Sample,StockSolution,"Biotin-XX, 1 mg/mL in DMSO"] if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
BiotinReagentVolume
The volume of BiotinReagent used to react with DetectionAntibodyVolume of the unconjugated CustomDetectionAntibody sample to prepare biotinylated detection antibody sample through bioconjugation synthesis.
Default Calculation: When a customizable cartridge is used, BiotinReagentVolume option is automatically set using the concentration of BiotinReagent and the antibody concentration to achieve a 10:1 molar ratio between BiotinReagent and the unconjugated CustomDetectionAntibody.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyConjugationBuffer
The buffer solution in which the reaction between the unconjugated CustomDetectionAntibody sample and BiotinReagent happens to prepare biotinylated detection antibody sample through bioconjugation synthesis.
Default Calculation: DetectionAntibodyConjugationBuffer option is automatically set to Model[Sample,StockSolution,"Sodium bicarbonate working stock 75 mg/mL"] if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyConjugationBufferVolume
The volume of DetectionAntibodyConjugationBuffer used to mix with the unconjugated CustomDetectionAntibody sample and BiotinReagent to provide a buffered environment for the bioconjugation reaction.
Default Calculation: When a customizable cartridge is used, DetectionAntibodyConjugationBufferVolume option is automatically set to 1/9 of total volume of DetectionAntibodyVolume and BiotinReagentVolume.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyConjugationContainer
The container in which the unconjugated CustomDetectionAntibody sample, BiotinReagent and DetectionAntibodyConjugationBuffer react to allow bioconjugation synthesis.
Default Calculation: DetectionAntibodyConjugationContainer option is automatically selected base on the total conjugation reaction volume if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyConjugationTime
The amount of reaction time that the CustomDetectionAntibody and BiotinReagent are allowed to react before purification happens.
Default Calculation: DetectionAntibodyConjugationTime option is automatically set to 1 Hour if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, $MaxExperimentTime] | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyConjugationTemperature
The temperature at which the reaction between the unconjugated CustomDetectionAntibody and BiotinReagent is conducted.
Default Calculation: DetectionAntibodyConjugationTemperature option is automatically set to Ambient if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[-20*Celsius, 150*Celsius]) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyPurificationColumn
The desalting spin column used to adsorb the molecules that are smaller than its molecular weight cut-off (MWCO) on its resin bed to remove the free biotinylation reagent molecules in order to purify the synthesis product of the CustomDetectionAntibody bioconjugation reaction. The spin column is operated following the manufacturer's instruction (Figure 3.5 in ExperimentCapillaryELISA help file).
Default Calculation: DetectionAntibodyPurificationColumn is automatically set to a 40K molecular weight cut-off Zeba desalting spin column depending on the total volume of the synthesis reaction, if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container, Vessel, Filter] or Object[Container, Vessel, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Vessel, Filter], Object[Container, Vessel, Filter]}] | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyColumnWashBuffer
The buffer solution loaded into the DetectionAntibodyPurificationColumn after its storage buffer is removed and before the sample is loaded. The DetectionAntibodyColumnWashBuffer flushed through DetectionAntibodyPurificationColumn to remove any residues from the spin column storage solution. The washing process is repeated 3 times to equilibrate the resin bed before sample loading.
Default Calculation: DetectionAntibodyColumnWashBuffer option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyConjugationStorageCondition
Indicates the condition under which the unused portion of the synthesized and purified biotinylated detection antibody sample is stored after the protocol is completed.
Default Calculation: DetectionAntibodyConjugationStorageCondition option is automatically set to Refrigerator if DetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyDilution
Indicates if biotinylated detection antibody dilution is required before loading into capillary ELISA cartridge.
Default Calculation: When a Customizable capillary ELISA cartridge is used, DetectionAntibodyDilution is automatically set to True if DetectionAntibodyResuspension and/or DetectionAntibodyConjugation is True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyTargetConcentration
The desired concentration of biotinylated detection antibody after dilution. The initial mass concentration of biotinylated detection antibody sample is acquired from the composition of the liquid state CustomDetectionAntibody sample or the DetectionAntibodyResuspension of the solid state CustomDetectionAntibody sample. A 50% concentration loss is factored in if conjugation is performed in the experiment.
Default Calculation: DetectionAntibodyTargetConcentration is automatically set to 3.5 Microgram/Milliliter if DetectionAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyDiluent
The buffer solution used to mix with the concentrated biotinylated detection antibody sample to achieve the desired DetectionAntibodyTargetConcentration for cartridge loading.
Default Calculation: DetectionAntibodyDiluent option is automatically set to Model[Sample, "Simple Plex Reagent Diluent"] if DetectionAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: experiment samples
Standard Curve
Standard
The standard samples with known concentration(s) of the analyte(s) that are used to make a dilution series and create a standard curve in the capillary ELISA experiment.
Default Calculation: If a customizable cartridge is used, Standard option is automatically set to the DefaultSampleModel of the Analytes.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
Standard Preparation
StandardResuspension
Indicates if resuspension is required to be performed on the solid state Standard sample to turn the sample into solution. If set to True, the original container of the Standard sample is centrifuged at 3000 RPM for 1 Minute to eliminate sample loss upon cap opening. Then the StandardResuspensionDiluent is delivered into the Standard's container and thorough mixing is achieved by inversion for 30 times.
Default Calculation: StandardResuspension is automatically set to True for a solid state Standard sample.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardResuspensionConcentration
The target concentration of the Standard sample in the solution after resuspension. The amount of the solid state Standard is used to calculate the volume of StandardResuspensionDiluent needed.
Default Calculation: If a pre-loaded cartridge is used in this protocol and the standard sample is for one of its analytes, StandardResuspensionConcentration is automatically set to 10 times of the analyte's upper limit of quantitation (ULOQ) provided by the assay developer. This information can be found in the corresponding Object[ManufacturingSpecification,CapillaryELISACartridge] of the analyte. Otherwise, StandardResuspension is automatically set to 100 Microgram/Milliliter if StandardResuspension is True.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: Standard
StandardResuspensionDiluent
The resuspension buffer mixed with the solid state Standard sample to dissolve the sample into solution.
Default Calculation: If a pre-loaded cartridge is used in this protocol and the standard sample is for one of its analytes, StandardResuspensionDiluent is automatically set to the recommended diluent for this analyte suggested by the assay developer. This information can be found in the corresponding Object[ManufacturingSpecification,CapillaryELISACartridge] of the analyte. Otherwise it is automatically set to Model[Sample,"Simple Plex Sample Diluent 13"] if StandardResuspension is True.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDilutionCurve
The collection of dilutions performed on the liquid-phase Standard sample. Dilution is required on all the loading samples for the best performance of microfluidics and the ELISA experiment is performed on all the diluted samples. Data from all measurements of all dilutions from a standard sample are used for the preparation of standard curve. For FixedDilutionVolume dilutions, the SampleAmount is the volume of the standard sample that is mixed with the DiluentVolume of the StandardDiluent. For FixedDilutionFactor dilutions, the AssayVolume is the volume of the sample that is created after being diluted by the specified DilutionFactor. For a pre-loaded capillary ELISA cartridge, please refer to Object[ManufacturingSpecification,CapillaryELISACartridge] of the analytes for their recommended minimum dilution factors. For a customizable cartridge, a minimum dilution factor of 0.5 is suggested.
Default Calculation: If Standard option is not Null and StandardSerialDilutionCurve is Null, StandardDilutionCurve is automatically set to {{60 Microliter,0.1},{60 Microliter,0.01},{60 Microliter,0.001},{60 Microliter,0.0001},{60 Microliter,0.00001}} to prepare a 60 Microliter dilution series of 5 orders.
Pattern Description: Fixed Dilution Factor or Fixed Dilution Volume or Null.
Programmatic Pattern: (({{GreaterEqualP[0*Microliter], GreaterEqualP[0*Microliter]}..} | {{GreaterEqualP[30*Microliter], RangeP[0, 1]}..}) | Automatic) | Null
Index Matches to: Standard
StandardSerialDilutionCurve
The collection of stepwise dilutions performed on the liquid-phase Standard sample. Dilution is required on all the loading samples for the best performance of microfluidics and the ELISA experiment is performed on all the diluted samples. Data from all measurements of all dilutions from a standard sample are used for the preparation of standard curve. To use non-diluted input sample, the first dilution can be set to a dilution factor of 1. For SerialDilutionVolumes, the TransferVolume is taken out of the sample and added to a second well with the DiluentVolume of the Diluent. After thorough mixing, the TransferVolume is taken out of that well and added to a third well. This is repeated to make NumberofDilutions diluted samples. For SerialDilutionFactors, the sample is diluted by the dilution factor at each transfer step. For a pre-loaded capillary ELISA cartridge, please refer to Object[ManufacturingSpecification,CapillaryELISACartridge] of the analytes for their recommended minimum dilution factors. For a customizable cartridge, a minimum dilution factor of 0.5 is suggested.
Default Calculation: If Standard option is uploaded and StandardDilutionCurve is uploaded, StandardSerialDilutionCurve option is automatically set to 5 NumberOfDilutions with ConstantDilutionFactor of 0.1 to generate a AssayVolume of 60 Microliter.
Pattern Description: Serial Dilution Factor or Serial Dilution Volumes or Null.
Programmatic Pattern: (({GreaterP[0*Microliter], GreaterEqualP[30*Microliter], GreaterP[1, 1]} | {GreaterEqualP[30*Microliter], {RangeP[0, 1], GreaterP[1, 1]} | {RangeP[0, 1]..}}) | Automatic) | Null
Index Matches to: Standard
StandardDiluent
The diluent buffer used to mixed with the Standard sample in order to make StandardDilutionCurve or StandardSerialDilutionCurve. Dilution with the recommended diluent is required on all the loading samples for the best performance of microfluidics.
Default Calculation: If a pre-loaded cartridge is used for the experiment, StandardDiluent option is automatically set to the recommended diluent provided by the assay developer for the specified Analytes or Cartridge. For a customizable cartridge, StandardDiluent option is set to Model[Sample,"Simple Plex Sample Diluent 13"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDilutionMixVolume
The volume that is pipetted up and down in the diluted standard sample to mix each sample thoroughly with StandardDiluent to make StandardDilutionCurve or StandardSerialDilutionCurve.
Default Calculation: StandardDilutionMixVolume is automatically set to 20 Microliter if Standard is not Null.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardDilutionNumberOfMixes
The number of pipette up and down cycles that is used to mix each standard sample thoroughly with StandardDiluent to make StandardDilutionCurve or StandardSerialDilutionCurve.
Default Calculation: StandardDilutionNumberOfMixes option is automatically set to 5 if Standard is not Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 20 in increments of 1 or Null.
Programmatic Pattern: (RangeP[0, 20, 1] | Automatic) | Null
Index Matches to: Standard
StandardDilutionMixRate
The speed at which the StandardDilutionMixVolume is pipetted up and down in the diluted standard sample to mix each sample thoroughly with StandardDiluent to make StandardDilutionCurve or StandardSerialDilutionCurve.
Default Calculation: StandardDilutionMixRate option is automatically set to 100 Microliter/Second if Standard is not Null.
Pattern Description: Greater than or equal to 0.4 microliters per second and less than or equal to 250 microliters per second or Null.
Programmatic Pattern: (RangeP[0.4*(Microliter/Second), 250*(Microliter/Second)] | Automatic) | Null
Index Matches to: Standard
Standard Capture Antibody Preparation
StandardCaptureAntibody
The standard capture antibody sample used in the sandwich ELISA experiment to bind with analytes in the standard sample through specific antigen-antibody interaction. The standard capture antibody must be labeled with digoxigenin, purified and diluted to preferably 3.5 Microgram/Milliliter before loading into the capillary ELISA cartridge. During the experiment, digoxigenin-labeled standard capture antibody sample first flows in the fluidic circuit to allow its binding to the immobilized anti-digoxigenin monoclonal antibody in the capillaries. Then the diluted standard samples are applied to allow the binding to standard capture antibody for immobilization. The provided StandardCaptureAntibody can be resuspended into solution, bioconjugated with digoxigenin and diluted as desired using the related options. Please note that if the same capture antibody preparation options are specified for multiple samples (standard samples and/or input samples), the preparation of digoxigenin-labeled capture antibody is performed together. Please consider using ExperimentSamplePreparation for more customized standard capture antibody preparation.
Default Calculation: If a customizable cartridge is used, StandardCaptureAntibody is automatically set based on the presented analyte (from Analytes option) in the standard sample. The Antibodies or SecondaryAntibodies field in Model[Molecule] of the analyte is inspected. If available, the DefaultSampleModel of a digoxigenin-labeled antibody is automatically selected as StandardCaptureAntibody. If no digoxigenin-labeled antibody is available, the DefaultSampleModel of an antibody with different epitopes with StandardDetectionAntibody (if uploaded) is automatically selected as StandardCaptureAntibody. If neither StandardCaptureAntibody or StandardDetectionAntibody has been specified and two antibodies with different epitopes are available for the analyte, the DefaultSampleModel of a polyclonal antibody is preferably selected for StandardCaptureAntibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyResuspension
Indicates if the StandardCaptureAntibody should be reconstituted with StandardCaptureAntibodyResuspensionDiluent. If set to True, the original container of the StandardCaptureAntibody sample is centrifuged at 3000 RPM for 1 Minute to eliminate sample loss upon cap opening. Then the StandardCaptureAntibodyResuspensionDiluent is delivered into the StandardCaptureAntibody's container and thorough mixing is achieved by inversion for 30 times.
Default Calculation: StandardCaptureAntibodyResuspension is automatically set to True for a solid state StandardCaptureAntibody sample in a customizable cartridge.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyResuspensionConcentration
The target final concentration of the resuspended StandardCaptureAntibody sample.
Default Calculation: StandardCaptureAntibodyResuspensionConcentration option is automatically set to 1 Milligram/Milliliter if StandardCaptureAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyResuspensionDiluent
The resuspension buffer mixed with the solid state StandardCaptureAntibody sample to dissolve the sample into solution.
Default Calculation: StandardCaptureAntibodyResuspensionDiluent option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if StandardCaptureAntibodyResuspension is True and a customizable cartridge is used. The amount of the solid state StandardCaptureAntibody sample is used to calculate the amount of StandardCaptureAntibodyResuspensionDiluent needed.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyConjugation
Indicates if bioconjugation reaction between digoxigenin NHS-ester and primary amines of the StandardCaptureAntibody is required to be performed to prepare digoxigenin-labeled standard capture antibody sample for capillary ELISA experiment. If set to True, the conjugation mixture is vortexed at 1900 RPM for 1 Minute to allow thorough mixing and then incubated on a heat block at StandardCaptureAntibodyConjugationTemperature for StandardCaptureAntibodyConjugationTime to allow complete reaction.
Default Calculation: StandardCaptureAntibodyConjugation is automatically set to True if the antibody composition of the StandardCaptureAntibody in the Customizable capillary ELISA cartridge does not show Model[Molecule,Protein,Antibody,"Anti-Digoxigenin Antibody"] as its SecondaryAntibodies.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyVolume
The volume of the unconjugated StandardCaptureAntibody sample used to react with StandardDigoxigeninReagent to prepare digoxigenin-labeled standard capture antibody sample through bioconjugation synthesis.
Default Calculation: StandardCaptureAntibodyVolume option is automatically set to 100 Microliter if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardDigoxigeninReagent
The digoxigenin NHS-ester reagent used to react with the primary amines of the unconjugated StandardCaptureAntibody sample to prepare digoxigenin-labeled standard capture antibody sample through bioconjugation synthesis.
Default Calculation: StandardDigoxigeninReagent option is automatically set to Model[Sample,StockSolution,"Digoxigenin-NHS, 0.67 mg/mL in DMF"] if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDigoxigeninReagentVolume
The volume of StandardDigoxigeninReagent used to react with StandardCaptureAntibodyVolume of the unconjugated StandardCaptureAntibody sample to prepare digoxigenin-labeled capture antibody sample through bioconjugation synthesis.
Default Calculation: When a customizable cartridge is used, StandardDigoxigeninReagentVolume option is automatically set using the concentration of StandardDigoxigeninReagent and the antibody concentration to achieve a 5:1 molar ratio between StandardDigoxigeninReagent and the unconjugated StandardCaptureAntibody.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyConjugationBuffer
The buffer solution in which the reaction between the unconjugated StandardCaptureAntibody sample and StandardDigoxigeninReagent happens to prepare digoxigenin-labeled capture antibody sample through bioconjugation synthesis.
Default Calculation: StandardCaptureAntibodyConjugationBuffer option is automatically set to Model[Sample,StockSolution,"Sodium bicarbonate working stock 75 mg/mL"] if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyConjugationBufferVolume
The volume of StandardCaptureAntibodyConjugationBuffer used to mix with the unconjugated StandardCaptureAntibody sample and StandardDigoxigeninReagent to provide a buffered environment for the bioconjugation reaction.
Default Calculation: When a customizable cartridge is used, StandardCaptureAntibodyConjugationBufferVolume option is automatically set to 1/9 of total volume of StandardCaptureAntibodyVolume and StandardDigoxigeninReagentVolume.
Pattern Description: Greater than or equal to 0 microliters or Null.
Programmatic Pattern: (GreaterEqualP[0*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyConjugationContainer
The container in which the unconjugated StandardCaptureAntibody sample, StandardDigoxigeninReagent and StandardCaptureAntibodyConjugationBuffer react to allow bioconjugation synthesis.
Default Calculation: StandardCaptureAntibodyConjugationContainer option is automatically selected base on the total conjugation reaction volume if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyConjugationTime
The amount of reaction time that the StandardCaptureAntibody and StandardDigoxigeninReagent are allowed to react before purification happens.
Default Calculation: StandardCaptureAntibodyConjugationTime option is automatically set to 1 Hour if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, $MaxExperimentTime] | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyConjugationTemperature
The temperature at which the reaction between the unconjugated StandardCaptureAntibody and StandardDigoxigeninReagent is conducted.
Default Calculation: StandardCaptureAntibodyConjugationTemperature option is automatically set to Ambient if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[-20*Celsius, 150*Celsius]) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyPurificationColumn
The desalting spin column used to adsorb the molecules that are smaller than its molecular weight cut-off (MWCO) on its resin bed to remove the free digoxigenin NHS-ester molecules in order to purify the synthesis product of the StandardCaptureAntibody bioconjugation reaction. The spin column is operated following the manufacturer's instruction (Figure 3.6 in ExperimentCapillaryELISA help file).
Default Calculation: StandardCaptureAntibodyPurificationColumn is automatically set to a 40K molecular weight cut-off Zeba desalting spin column depending on the total volume of the synthesis reaction, if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container, Vessel, Filter] or Object[Container, Vessel, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Vessel, Filter], Object[Container, Vessel, Filter]}] | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyColumnWashBuffer
The buffer solution loaded into the StandardCaptureAntibodyPurificationColumn after its storage buffer is removed and before the sample is loaded. The StandardCaptureAntibodyColumnWashBuffer flushed through StandardCaptureAntibodyPurificationColumn to remove any residues from the spin column storage solution. The washing process is repeated 3 times to equilibrate the resin bed before sample loading.
Default Calculation: StandardCaptureAntibodyColumnWashBuffer option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyConjugationStorageCondition
Indicates the condition under which the unused portion of the synthesized and purified digoxigenin-labeled standard capture antibody sample is stored after the protocol is completed.
Default Calculation: StandardCaptureAntibodyConjugationStorageCondition option is automatically set to Refrigerator if StandardCaptureAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyDilution
Indicates if digoxigenin-labeled standard capture antibody dilution is required before loading into capillary ELISA cartridge.
Default Calculation: When a Customizable capillary ELISA cartridge is used, StandardCaptureAntibodyDilution is automatically set to True if StandardCaptureAntibodyResuspension and/or StandardCaptureAntibodyConjugation is True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyTargetConcentration
The desired concentration of digoxigenin-labeled standard capture antibody after dilution. The initial mass concentration of digoxigenin-labeled standard capture antibody sample is acquired from the composition of the liquid state StandardCaptureAntibody sample or the StandardCaptureAntibodyResuspension of the solid state StandardCaptureAntibody sample. A 50% concentration loss is factored in if conjugation is performed in the experiment.
Default Calculation: StandardCaptureAntibodyTargetConcentration is automatically set to 3.5 Microgram/Milliliter if StandardCaptureAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyDiluent
The buffer solution used to mix with the concentrated digoxigenin-labeled standard capture antibody sample to achieve the desired StandardCaptureAntibodyTargetConcentration for cartridge loading.
Default Calculation: StandardCaptureAntibodyDiluent option is automatically set to Model[Sample, "Simple Plex Reagent Diluent"] if StandardCaptureAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
Standard Detection Antibody Preparation
StandardDetectionAntibody
The standard detection antibody sample used in the sandwich ELISA experiment. After immobilization of the analytes in the microfluidic channels, the standard detection antibodies bind to analyte epitopes distinct from the standard capture antibodies through antigen-antibody interaction. Then the streptavidin-conjugated fluorescent dyes can be attached through biotin-streptavidin interaction for data acquiring. The standard detection antibody must be modified with biotin, purified and diluted to preferably 3.5 Microgram/Milliliter before loading into the capillary ELISA cartridge. The provided StandardDetectionAntibody can be resuspended into solution, bioconjugated with biotin and diluted as desired using the related options. Please note that if the same detection antibody preparation options are specified for multiple samples (standard samples and/or input samples), the preparation of biotinylated detection antibody is performed together. Please consider using ExperimentSamplePreparation for more customized standard detection antibody preparation.
Default Calculation: If a customizable cartridge is used, StandardDetectionAntibody is automatically set based on the presented analyte (from Analytes option) in the standard sample. The Antibodies or SecondaryAntibodies field in Model[Molecule] of the main analyte is inspected. If available, the DefaultSampleModel of a biotinylated antibody is automatically selected as StandardDetectionAntibody. If no biotinylated antibody is available, the DefaultSampleModel of an antibody with different epitopes with StandardCaptureAntibody (if specified) is automatically selected as CustomDetectionAntibody. If neither StandardCaptureAntibody or StandardDetectionAntibody has been specified and two antibodies with different epitopes are available for the analyte, the DefaultSampleModel of a monoclonal antibody is preferably selected for StandardDetectionAntibody.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyResuspension
Indicates if the StandardCaptureAntibody should be reconstituted with StandardCaptureAntibodyResuspensionDiluent. If set to True, the original container of the StandardDetectionAntibody sample is centrifuged at 3000 RPM for 1 Minute to eliminate sample loss upon cap opening. Then the StandardDetectionAntibodyResuspensionDiluent is delivered into the StandardDetectionAntibody's container and thorough mixing is achieved by inversion for 30 times.
Default Calculation: StandardDetectionAntibodyResuspension is automatically set to True for a solid state StandardDetectionAntibody sample in a Customizable capillary ELISA cartridge.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyResuspensionConcentration
The target final concentration of the resuspended StandardDetectionAntibody sample.
Default Calculation: StandardDetectionAntibodyResuspensionConcentration option is automatically set to 1 Milligram/Milliliter if StandardDetectionAntibodyResuspension is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyResuspensionDiluent
The resuspension buffer mixed with the solid state StandardDetectionAntibody sample to dissolve the sample into solution.
Default Calculation: StandardDetectionAntibodyResuspensionDiluent option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if StandardDetectionAntibodyResuspension is True and a customizable cartridge is used. The amount of the solid state StandardDetectionAntibody sample is used to calculate the amount of StandardDetectionAntibodyResuspensionDiluent needed.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyConjugation
Indicates if bioconjugation reaction between Biotin-XX-SE (6-((6-((Biotinoyl)Amino)Hexanoyl)Amino)Hexanoic Acid, Succinimidyl Ester) and primary amines of the StandardDetectionAntibody is required to be performed to prepare biotinylated detection antibody sample for capillary ELISA experiment. If set to True, the conjugation mixture is vortexed at 1900 RPM for 1 Minute to allow thorough mixing and then incubated on a heat block at StandardDetectionAntibodyConjugationTemperature for StandardDetectionAntibodyConjugationTime to allow complete reaction.
Default Calculation: StandardDetectionAntibodyConjugation is automatically set to True if the antibody composition of the StandardDetectionAntibody in the Customizable capillary ELISA cartridge does not show Model[Molecule,Protein,"Streptavidin"] as its Targets.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyVolume
The volume of the unconjugated standard detection antibody sample used to react with StandardBiotinReagent to prepare biotinylated standard detection antibody sample through bioconjugation synthesis.
Default Calculation: StandardDetectionAntibodyVolume option is automatically set to 100 Microliter if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardBiotinReagent
The Biotin-XX-SE (6-((6-((Biotinoyl)Amino)Hexanoyl)Amino)Hexanoic Acid, Succinimidyl Ester) or other biotinylation reagent used to mix with the unconjugated StandardDetectionAntibody sample to prepare biotinylated standard detection antibody sample through bioconjugation synthesis.
Default Calculation: StandardBiotinReagent option is automatically set to Model[Sample,StockSolution,"Biotin-XX, 1 mg/mL in DMSO"] if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardBiotinReagentVolume
The volume of StandardBiotinReagent used to react with StandardDetectionAntibodyVolume of the unconjugated StandardDetectionAntibody sample to prepare biotinylated detection antibody sample through bioconjugation synthesis.
Default Calculation: When a customizable cartridge is used, StandardBiotinReagentVolume option is automatically set using the concentration of StandardBiotinReagent and the antibody concentration to achieve a 10:1 molar ratio between StandardBiotinReagent and the unconjugated StandardDetectionAntibody.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyConjugationBuffer
The buffer solution in which the reaction between the unconjugated StandardDetectionAntibody sample and StandardBiotinReagent happens to prepare biotinylated standard detection antibody sample through bioconjugation synthesis.
Default Calculation: StandardDetectionAntibodyConjugationBuffer option is automatically set to Model[Sample,StockSolution,"Sodium bicarbonate working stock 75 mg/mL"] if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyConjugationBufferVolume
The volume of StandardDetectionAntibodyConjugationBuffer used to mix with the unconjugated StandardDetectionAntibody sample and StandardBiotinReagent to provide a buffered environment for the bioconjugation reaction.
Default Calculation: When a customizable cartridge is used, StandardDetectionAntibodyConjugationBufferVolume option is automatically set to 1/9 of total volume of StandardDetectionAntibodyVolume and StandardBiotinReagentVolume if both are uploaded.
Pattern Description: Greater than 0 microliters or Null.
Programmatic Pattern: (GreaterP[0*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyConjugationContainer
The container in which the unconjugated StandardDetectionAntibody sample, StandardBiotinReagent and StandardDetectionAntibodyConjugationBuffer react to allow bioconjugation synthesis.
Default Calculation: StandardDetectionAntibodyConjugationContainer option is automatically selected base on the total conjugation reaction volume if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyConjugationTime
The amount of reaction time that the StandardDetectionAntibody and StandardBiotinReagent are allowed to react before purification happens.
Default Calculation: StandardDetectionAntibodyConjugationTime option is automatically set to 1 Hour if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Greater than or equal to 0 minutes and less than or equal to 72 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, $MaxExperimentTime] | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyConjugationTemperature
The temperature at which the reaction between the unconjugated StandardDetectionAntibody and StandardBiotinReagent is conducted.
Default Calculation: StandardDetectionAntibodyConjugationTemperature option is automatically set to Ambient if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[-20*Celsius, 150*Celsius]) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyPurificationColumn
The desalting spin column used to adsorb the molecules that are smaller than its molecular weight cut-off (MWCO) on its resin bed to remove the free biotinylation reagent molecules in order to purify the synthesis product of the StandardDetectionAntibody bioconjugation reaction. The spin column is operated following the manufacturer's instruction (Figure 3.7 in ExperimentCapillaryELISA help file).
Default Calculation: StandardDetectionAntibodyPurificationColumn is automatically set to a 40K molecular weight cut-off Zeba desalting spin column depending on the total volume of the synthesis reaction, if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Container, Vessel, Filter] or Object[Container, Vessel, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Vessel, Filter], Object[Container, Vessel, Filter]}] | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyColumnWashBuffer
The buffer solution loaded into the StandardDetectionAntibodyPurificationColumn after its storage buffer is removed and before the sample is loaded. The StandardDetectionAntibodyColumnWashBuffer flushed through DetectionAntibodyPurificationColumn to remove any residues from the spin column storage solution. The washing process is repeated 3 times to equilibrate the resin bed before sample loading.
Default Calculation: StandardDetectionAntibodyColumnWashBuffer option is automatically set to Model[Sample, StockSolution, "Filtered PBS, Sterile"] if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyConjugationStorageCondition
Indicates the condition under which the unused portion of the synthesized and purified biotinylated detection antibody sample is stored after the protocol is completed.
Default Calculation: StandardDetectionAntibodyConjugationStorageCondition option is automatically set to Refrigerator if StandardDetectionAntibodyConjugation is True and a customizable cartridge is used.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting, Oven} or Disposal or Null.
Programmatic Pattern: ((SampleStorageTypeP | Disposal) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyDilution
Indicates if biotinylated standard detection antibody dilution is required before loading into capillary ELISA cartridge.
Default Calculation: When a Customizable capillary ELISA cartridge is used, StandardDetectionAntibodyDilution is automatically set to True if StandardDetectionAntibodyResuspension and/or StandardDetectionAntibodyConjugation is True.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyTargetConcentration
The desired concentration of biotinylated standard detection antibody after dilution. The initial mass concentration of biotinylated standard detection antibody sample is acquired from the composition of the liquid state StandardDetectionAntibody sample or the StandardDetectionAntibodyResuspension of the solid state StandardDetectionAntibody sample. A 50% concentration loss is factored in if conjugation is performed in the experiment.
Default Calculation: StandardDetectionAntibodyTargetConcentration is automatically set to 3.5 Microgram/Milliliter if StandardDetectionAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: Greater than 0 molar or greater than 0 grams per liter or Null.
Programmatic Pattern: ((GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)]) | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyDiluent
The buffer solution used to mix with the concentrated biotinylated detection antibody sample to achieve the specified StandardDetectionAntibodyTargetConcentration for cartridge loading.
Default Calculation: StandardDetectionAntibodyDiluent option is automatically set to Model[Sample, "Simple Plex Reagent Diluent"] if StandardDetectionAntibodyDilution is True and a customizable cartridge is used.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
Washing
WashBuffer
The buffer that is used to remove excess reagents between the antibody binding steps.
Default Value: Model[Sample, id:4pO6dM50p9kw]
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample.
Programmatic Pattern: ObjectP[{Model[Sample], Object[Sample]}] | _String
Cartridge Loading
LoadingVolume
The volume of each prepared input sample loaded into the capillary ELISA cartridge.
Default Value: 50 microliters
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 90 microliters.
Programmatic Pattern: RangeP[25*Microliter, 90*Microliter]
Index Matches to: experiment samples
CaptureAntibodyLoadingVolume
The volume of the digoxigenin-labeled capture antibody sample loaded into the capture antibody well of the customizable capillaryELISA cartridge plate.
Default Calculation: CaptureAntibodyLoadingVolume option is automatically set to 50 Microliter if a customizable cartridge is used.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: experiment samples
DetectionAntibodyLoadingVolume
The volume of the biotinylated detection antibody sample to be loaded into the detection antibody well of the customizable capillaryELISA cartridge plate.
Default Calculation: DetectionAntibodyLoadingVolume option is automatically set to 50 Microliter iif a customizable cartridge is used.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: experiment samples
StandardLoadingVolume
The volume of each prepared standard sample loaded into the capillary ELISA cartridge.
Default Calculation: StandardLoadingVolume option is automatically set to 50 Microliter if Standard option is not Null.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 90 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 90*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardCaptureAntibodyLoadingVolume
The volume of the digoxigenin-labeled standard capture antibody sample to be loaded into the capture antibody well of the customizable capillaryELISA cartridge plate.
Default Calculation: StandardCaptureAntibodyLoadingVolume option is automatically set to 50 Microliter if a customizable cartridge is used.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardDetectionAntibodyLoadingVolume
The volume of the biotinylated standard detection antibody sample to be loaded into the detection antibody well of the customizable capillaryELISA cartridge plate.
Default Calculation: StandardDetectionAntibodyLoadingVolume option is automatically set to 50 Microliter if a customizable cartridge is used.
Pattern Description: Greater than or equal to 25 microliters and less than or equal to 50 microliters or Null.
Programmatic Pattern: (RangeP[25*Microliter, 50*Microliter] | Automatic) | Null
Index Matches to: Standard
Data Processing
StandardComposition
The known concentration(s) of presented analyte(s) in the Standard sample before dilution. This information can be used to generate standard curve.
Default Calculation: KnownConcentration is automatically set to StandardResuspensionConcentration if the resuspension is performed within this protocol. Otherwise it is automatically set using the information from the Composition field of the Standard sample.
Pattern Description: List of one or more {Concentration, Analyte} entries or Null.
Programmatic Pattern: ({{GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)], ObjectP[{Model[Molecule]}]}..} | Automatic) | Null
Index Matches to: Standard
SpikeConcentration
The known concentration(s) of the analyte(s) in SpikeSample. This information is used for calculation of the analyte concentration(s) in unknown sample if a standard curve is available. For a pre-loaded cartridge with more than 1 analyte, concentrations of different analytes can be provided for the SpikeSample.
Default Calculation: SpikeConcentration is automatically set using the information from the Composition field of the SpikeSample.
Pattern Description: List of one or more {Concentration, Analyte} entries or Null.
Programmatic Pattern: ({{GreaterP[0*Molar] | GreaterP[0*(Gram/Liter)], ObjectP[{Model[Molecule]}]}..} | Automatic) | Null
Index Matches to: experiment samples
Model Input
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and when the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise set to Model[Container, Vessel, "2mL Tube"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: (ObjectP[Model[Container]] | Automatic) | Null
Index Matches to: experiment samples
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
Index Matches to: experiment samples