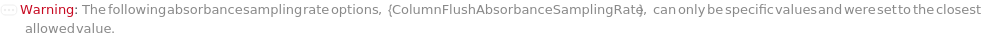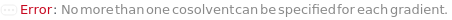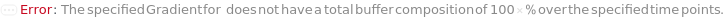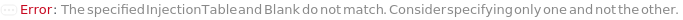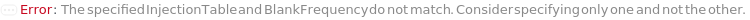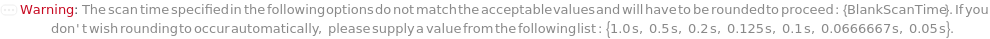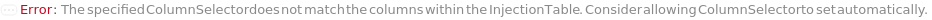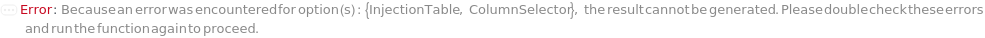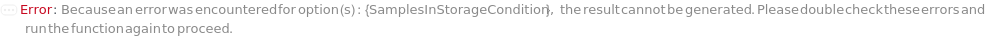ExperimentSupercriticalFluidChromatography
ExperimentSupercriticalFluidChromatography[Samples]⟹Protocol
generates a Protocol to separate and analyze Samples via Supercritical Fluid Chromatography (SFC).
Supercritical fluid chromatography (SFC) involves a pressured carbon dioxide stream, into which, samples are injected and flow through the stationary phase column. Analytes pass over resin within the column and selectively adsorb based on their relative affinity, leading to differential retention of unique analyte molecules. Molecules are carried downstream for chemical property analysis. On the whole, molecular components are separated into analyzable, quantifiable constituents. For instance, fat-soluble vitamins can be separated and quantified from a mixture. In contrast to high-performance liquid chromatography (HPLC), SFC is ideal for analytes with poor water solubility or for separating enantiomers from one another.
Experimental Principles
Figure 1.1: Procedural overview of a Supercritical Fluid Chromatography experiment. Step 1: During the prime system, the system is rinsed with cleaning solutions. Step 2: The stationary Column(s) are installed and equilibrated to measurement conditions. Step 3: Samples are then introduced into the flow path. Step 4: The analytes are selectively retained on the downstream Column. Step 5: Upon exit from the column, the now separated analytes are analyzed according to their physical property. Step 6: After final sample measurement, the Column(s) are rinsed, removed from the system, and stowed. Step 7: After the column is removed the system is rinsed with cleaning solution.
Instrumentation
Waters UPC2 PDA QDa
Figure 2.1.1: Supercritical carbon dioxide is steadily pumped through the instrument consisting of a 6-port valve system, adsorbent column, and detectors. Cosolvent can be supplemented into the fluid flow in order to change retention properties of the column. Samples within the Autosampler are loaded into the Sample loop via positive displacement from the Syringe. The rotation of the valve exposes the sample loop, thereby carrying the sample downstream to the column. Columns can be switched between injector via the column changer. Within the column, molecular constituents are separated by adsorption -- a function of the cosolvent composition in the carbon dioxide, cosolvent properties, additives in the cosolvent, column, sample properties and temperature. Column effluent is split between MassSpectrometry and PhotoDiodeArray detection. In the flow path to MassSpectrometry, MakeupSolvent can be mixed in to facilitate analyte ionization. Flow coming out the PhotoDiodeArray is pressurized with automatic back pressure regulation. This back pressure ensures that the carbon dioxide remains supercritical within the entire system. Furthermore, increasing back pressure will compress the carbon dioxide, changing the sample chromatography, and will divert more flow toward the mass spectrometry detection.
Figure 2.1.2: Principle of PhotodiodeArray detection. Filtered light (across a range of wavelengths) passes through the flow downstream of the column. Presence of light-absorbing molecules will result in less light transmission to the recipient Diode, thereby producing a chromatographic peak for each specific wavelength.
Figure 2.1.3: Principle of MassSpectrometry detection. Analytes flow into the source block and are evaporated under a stream of sheath gas. ESICapillaryVoltage is then applied to the capillary tubing tip to produce an electrospray, which turns into single gas-phase ions due to additional evaporation and ProbeTemperature. Ions then enter the mass spectrometer through the source cone and follow the ion path via an applied voltage (SamplingConeVoltage) to generate a focused ion beam. Ions travel through the quadrupole mass analyzer (Q1), where their mass is resolved by a specific mass-to-charge ratio (m/z, MassDetection). Spectra are continuously acquired for ions either inside MassDetection as a ranged value or as a fixed value, hitting the detector during ScanTime, and a total mass spectrum is generated.
Experiment Options
Protocol
Instrument
Pattern Description: An object of type or subtype Model[Instrument, SupercriticalFluidChromatography] or Object[Instrument, SupercriticalFluidChromatography]
Programmatic Pattern: ObjectP[{Model[Instrument, SupercriticalFluidChromatography], Object[Instrument, SupercriticalFluidChromatography]}]
Scale
Indicates if the experiment is intended purify or analyze material. Analytical indicates that specific measurements will be employed (e.g the absorbance of the flow with injected sample for a given wavelength) and is amendable for quantification.
SeparationMode
The category of separation to be used. This option is used to resolve the column, cosolvents, and column temperatures.
Detector
The type measurement to employ. Currently, we offer PhotoDiodeArray (measures the absorbance of a range of wavelengths) and MassSpectrometry (ionizes the analytes and measures the abundance of a given mass to charge ratio).
Pattern Description: List of one or more Temperature, Pressure, PhotoDiodeArray, or MassSpectrometry entries or Temperature, Pressure, PhotoDiodeArray, or MassSpectrometry.
Programmatic Pattern: (Temperature | Pressure | PhotoDiodeArray | MassSpectrometry) | {(Temperature | Pressure | PhotoDiodeArray | MassSpectrometry)..}
Column
Item containing the stationary phase through which the CO2, Cosolvent, and input samples flow. It adsorbs and separates the molecules within the sample based on the properties of the mobile phase, Samples, Column material, and ColumnTemperature.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
ColumnSelector
All the columns loaded into the Instrument's column selector and referenced in Column, StandardColumn, and BlankColumn options.
Default Calculation: Automatically set by running DeleteDuplicates on the Column, StandardColumn, and BlankColumn option.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
ColumnTemperature
Default Calculation: Automatically set from the temperature within the Gradient option; otherwise, set to 30 Celsius.
Pattern Description: Ambient or greater than or equal to 20 degrees Celsius and less than or equal to 90 degrees Celsius in increments of 1 degree Celsius or Null.
ColumnOrientation
The direction of the Column with respect to the flow. Forward indicates that the Column will be placed in the direction indicated by the column manufacturer for standard operation. Reverse indicates that the Column will be placed in the opposite direction indicated by the column manufacturer for standard operation and is typically used to clean the column.
Default Calculation: Automatically set to Forward for each column available; otherwise, set to Null.
NumberOfReplicates
The number of times to repeat measurements on each provided sample(s). If Aliquot -> True, this also indicates the number of times each provided sample will be aliquoted.
CosolventA
Default Calculation: Automatically set from SeparationMode or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
CosolventB
Default Calculation: Automatically set from SeparationMode or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
CosolventC
Default Calculation: Automatically set from SeparationMode or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
CosolventD
Default Calculation: Automatically set from SeparationMode or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Sample Parameters
InjectionTable
The order of sample, Standard, and Blank sample loading into the Instrument during measurement and/or collection. Also includes the flushing and priming of the column.
Default Calculation: Determined to the order of input samples articulated. Standard and Blank samples are inserted based on the determination of StandardFrequency and BlankFrequency. For example, StandardFrequency -> FirstAndLast and BlankFrequency -> Null result in Standard samples injected first, then samples, and then the Standard sample set again.
Pattern Description: List of one or more {Type, Sample, InjectionVolume, Column, Gradient} or {Type, Sample, InjectionVolume, Column, Gradient} entries.
Programmatic Pattern: {({Standard | Sample | Blank, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic, RangeP[0*Microliter, 50*Microliter, 1*Microliter] | Automatic, ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic, ObjectP[Object[Method, SupercriticalFluidGradient]] | Automatic} | {ColumnPrime | ColumnFlush, Automatic | Null, Automatic | Null, ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic, ObjectP[Object[Method, SupercriticalFluidGradient]] | Automatic})..} | Automatic
SampleTemperature
The temperature at which the samples, Standard, and Blank are kept in the instrument's autosampler prior to injection on the column.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 40 degrees Celsius in increments of 1 degree Celsius.
InjectionVolume
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 50 microliters.
NeedleWashSolution
Gradient
CO2Gradient
The composition of the supercritical CO2 within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for CO2Gradient->{{0 Minute, 90 Percent},{30 Minute, 100 Percent}}, the percentage of CO2Gradient in the flow will rise such that at 15 minutes, the composition should be 95*Percent.
Default Calculation: Automatically set from Gradient option or implicitly set from GradientA, GradientB, GradientC, and GradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, CO2 Composition} entries.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic
GradientA
The composition of CosolventA within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientA->{{0 Minute, 0 Percent},{30 Minute, 10 Percent}}, the percentage of CosolventA in the flow will rise such that at 15 minutes, the composition should be 5*Percent.
Default Calculation: Automatically set from Gradient option or implicitly set from CO2Gradient, GradientB, GradientC, and GradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
GradientB
The composition of CosolventB within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientB->{{0 Minute, 0 Percent},{30 Minute, 10 Percent}}, the percentage of CosolventB in the flow will rise such that at 15 minutes, the composition should be 5*Percent.
Default Calculation: Automatically set from Gradient option or implicitly set from CO2Gradient, GradientA, GradientC, and GradientD options. If no other gradient options are specified, will default to 0 Percent for the run.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
GradientC
The composition of CosolventC within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientC->{{0 Minute, 0 Percent},{30 Minute, 10 Percent}}, the percentage of CosolventC in the flow will rise such that at 15 minutes, the composition should be 5*Percent.
Default Calculation: Automatically set from Gradient option or implicitly set from CO2Gradient, GradientA, GradientB, and GradientD options. If no other gradient options are specified, will default to 0 Percent for the run.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
GradientD
The composition of CosolventD within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientD->{{0 Minute, 0 Percent},{30 Minute, 10 Percent}}, the percentage of CosolventD in the flow will rise such that at 15 minutes, the composition should be 5*Percent.
Default Calculation: Automatically set from Gradient option or implicitly set from CO2Gradient, GradientA, GradientB, and GradientC options. If no other gradient options are specified, will default to 0 Percent for the run.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
FlowRate
The speed of the fluid through the pump. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: Automatically set from Type and Scale or inherited from the method given in the Gradient option.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 4 milliliters per minute or list of one or more {Time, Flow Rate} entries.
Programmatic Pattern: (RangeP[0*(Milliliter/Minute), 4*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], RangeP[0*(Milliliter/Minute), 4*(Milliliter/Minute)]}..}) | Automatic
GradientStart
A shorthand option to specify the starting CosolventA composition in the fluid flow. This option must be specified with GradientEnd and GradientDuration.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
GradientEnd
A shorthand option to specify the final CosolventA composition in the fluid flow. This option must be specified with GradientStart and GradientDuration.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
GradientDuration
EquilibrationTime
A shorthand option to specify the duration of equilibration at the starting buffer composition at the start of a GradientA.
FlushTime
A shorthand option to specify the duration of equilibration at the final buffer composition at the end of a GradientA.
BackPressure
The pressure differential between the outlet of the system and the atmosphere. Higher BackPressure will increase the density of the supercritical CO2 mobile phase, thereby affecting analyte retention.
Furthermore, higher BackPressure increases flow rate of the column effluent into the mass spectrometer. BackPressure can be changed at different timepoints during the measurement; however, unlike CO2Gradient and FlowRate, BackPressure is not linearly interpolated and, rather, changes stepwise.
Default Calculation: Automatically set to specification within Gradient option. If Gradient option is not specified, defaults to 1800*PSI.
Pattern Description: Greater than or equal to 1000 pounds‐force per inch squared and less than or equal to 6000 pounds‐force per inch squared or list of one or more {Time, BackPressure} entries.
Programmatic Pattern: (RangeP[1000*PSI, 6000*PSI] | {{GreaterEqualP[0*Minute], RangeP[1000*PSI, 6000*PSI]}..}) | Automatic
Gradient
The composition over time in the fluid flow. Specific parameters of an object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the Gradient options (e.g. CO2Gradient, GradientA, GradientB, GradientC, GradientD, FlowRate, GradientStart, GradientEnd, GradientDuration, EquilibrateTime, BackPressure).
Pattern Description: An object of type or subtype Object[Method, SupercriticalFluidGradient] or list of one or more {Time, CO2 Composition, Cosolvent A Composition, Cosolvent B Composition, Cosolvent C Composition, Cosolvent D Composition, Back Pressure, Flow Rate} entries.
Programmatic Pattern: (ObjectP[Object[Method, SupercriticalFluidGradient]] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], GreaterEqualP[0*PSI], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic
General
MaxAcceleration
Pattern Description: Greater than or equal to 0.067 milliliters per minute squared and less than or equal to 200 milliliters per minute squared or Null.
Programmatic Pattern: RangeP[0.067*(Milliliter/Minute/Minute), 200*(Milliliter/Minute/Minute)] | Null
Detector Parameters
IonMode
Default Calculation: Automatically set to positive ion mode, unless the sample is acidic (pH<=5 or pKa<=8).
MakeupSolvent
The buffer mixed with column effluent in flow path to mass spectrometer. Facilitates molecular ionization.
Default Value: Model[Sample, StockSolution, 90% Methanol with 0.1% formic acid and 0.1% ammonium acetate]
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Calibrant
A sample with components of known mass-to-charge ratios (m/z) used to calibrate the mass spectrometer. In the chosen ion polarity mode, the calibrant should contain at least 3 masses spread over the mass range of interest.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
MakeupFlowRate
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 1 milliliter per minute.
MassDetection
Programmatic Pattern: {RangeP[30*(Gram/Mole), 1200*(Gram/Mole), 1*(Gram/Mole)]..} | All | RangeP[30*(Gram/Mole), 1199*(Gram/Mole), 1*(Gram/Mole)] ;; RangeP[31*(Gram/Mole), 1200*(Gram/Mole), 1*(Gram/Mole)]
ScanTime
The duration of time allowed to pass between each spectral acquisition. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired during the measurement.
ProbeTemperature
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius in increments of 1 degree Celsius.
ESICapillaryVoltage
The applied voltage differential between the injector and the inlet plate for the mass spectrometry in order to ionize analyte molecules.
Pattern Description: Greater than or equal to 0.3 kilovolts and less than or equal to 1.5 kilovolts.
SamplingConeVoltage
The voltage differential between ion spray entry and the quadrupole mass analyzer. SamplingConeVoltage facilitates ionization of the molecules.
This voltage normally optimizes between 25 and 100 V and should be adjusted for sensitivity depending on compound and charge state.
For multiply charged species it is typically set to to 40-50 V, and higher for singly charged species. SamplingConeVoltage can be specified as constant value for all m/z species or
as a ramp that linearly maps to the range specified for MassDetection. For example, if MassDetection is 100 to 1000 Da, and SamplingConeVoltage is 10 to 100 Volt, then 50 volts will be
applied to ions with m/z of 500 Da. The ramp option is only available when MassDetection is a range.
Programmatic Pattern: RangeP[0.1*Volt, 100*Volt] | RangeP[0.1*Volt, 100*Volt] ;; RangeP[0.1*Volt, 100*Volt]
MassDetectionGain
AbsorbanceWavelength
Programmatic Pattern: (RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 790*Nanometer, 1*Nanometer] ;; RangeP[200*Nanometer, 800*Nanometer, 1*Nanometer]) | Null
WavelengthResolution
The increment of wavelength for the range of light passed through the flow for absorbance measurement with the PDA detector.
Default Calculation: Automatically set to 1.2*Nanometer if AbsorbanceWavelength is a range; otherwise, set to Null.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
UVFilter
Indicates whether or not to block UV wavelengths (less than 210 nm) from being transmitted through the sample for PhotoDiodeArray detectors.
AbsorbanceSamplingRate
Indicates the frequency of absorbance measurement. Lower values will be less susceptible to noise but will record less frequently across time.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second.
Standard
Standard
A reference compound to inject to the instrument, often used for quantification or to check internal measurement consistency.
Default Calculation: Automatically set from SeparationMode when any other Standard option is specified, otherwise set to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
StandardInjectionVolume
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 50 microliters or Null.
StandardFrequency
Specify the frequency at which Standard measurements will be inserted in the measurement sequence. If specified to a number, indicates how often the Standard measurements will run among the analyte samples; for example, if 5 input samples are measured and StandardFrequency is 2, then Standard measurement will occur after the first two samples and again after the third and fourth.
Pattern Description: Greater than 0 in increments of 1 or None, First, Last, FirstAndLast, or GradientChange or Null.
Programmatic Pattern: (((None | First | Last | FirstAndLast | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
StandardColumn
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or list of one or more an object of type or subtype Model[Item, Column] or Object[Item, Column] entries or Null.
Programmatic Pattern: (({ObjectP[{Model[Item, Column], Object[Item, Column]}]..} | ObjectP[{Model[Item, Column], Object[Item, Column]}]) | Automatic) | Null
StandardColumnTemperature
Default Calculation: Automatically set from ColumnTemperature. Automatic resolution can be inherited from the StandardGradient option.
Pattern Description: Ambient or greater than or equal to 20 degrees Celsius and less than or equal to 90 degrees Celsius in increments of 1 degree Celsius or Null.
StandardCO2Gradient
The composition of the supercritical CO2 within the flow, defined for specific time points for standard samples.
Default Calculation: Automatically set from StandardGradient option or implicitly set from StandardGradientA, StandardGradientB, StandardGradientC, and StandardGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, CO2 Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
StandardGradientA
The composition of CosolventA within the flow, defined for specific time points for Standard measurement.
Default Calculation: Automatically set from StandardGradient option or implicitly set from StandardCO2Gradient, StandardGradientB, StandardGradientC, and StandardGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
StandardGradientB
The composition of CosolventB within the flow, defined for specific time points for Standard measurement.
Default Calculation: Automatically set from StandardGradient option or implicitly set from StandardCO2Gradient, StandardGradientA, StandardGradientC, and StandardGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
StandardGradientC
The composition of CosolventC within the flow, defined for specific time points for Standard measurement.
Default Calculation: Automatically set from StandardGradient option or implicitly set from StandardCO2Gradient, StandardGradientA, StandardGradientB, and StandardGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
StandardGradientD
The composition of CosolventD within the flow, defined for specific time points for Standard measurement.
Default Calculation: Automatically set from StandardGradient option or implicitly set from StandardCO2Gradient, StandardGradientA, StandardGradientB, and StandardGradientC options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
StandardFlowRate
Default Calculation: Automatically set from SeparationMode and Scale or inherited from the method given in the StandardGradient option.
Pattern Description: Greater than or equal to 0 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
StandardBackPressure
The pressure differential between the outlet of the system and the atmosphere for Standard sample measurement.
StandardBackPressure can be changed at different timepoints during the measurement; however, unlike StandardCO2Gradient and StandardFlowRate, StandardBackPressure is not linearly interpolated and, rather, changes stepwise.
Default Calculation: Automatically set to specification within Gradient option; otherwise, set to the same as the first entry in BackPressure.
Pattern Description: Greater than or equal to 1000 pounds‐force per inch squared and less than or equal to 6000 pounds‐force per inch squared or list of one or more {Time, BackPressure} entries or Null.
Programmatic Pattern: ((RangeP[1000*PSI, 6000*PSI] | {{GreaterEqualP[0*Minute], RangeP[1000*PSI, 6000*PSI]}..}) | Automatic) | Null
StandardGradient
The composition over time in the fluid flow for Standard samples. Specific parameters of an object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the Gradient options (e.g. StandardCO2Gradient, StandardGradientA, StandardGradientB, StandardGradientC, StandardGradientD, StandardFlowRate, GradientDuration, StandardBackPressure).
Pattern Description: An object of type or subtype Object[Method, SupercriticalFluidGradient] or list of one or more {Time, CO2 Composition, Cosolvent A Composition, Cosolvent B Composition, Cosolvent C Composition, Cosolvent D Composition, Back Pressure, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, SupercriticalFluidGradient]] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], GreaterEqualP[0*PSI], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
StandardGradientDuration
StandardIonMode
StandardMakeupFlowRate
The speed of the MakeupSolvent to supplement column effluent for the mass spectrometry measurement of Standard samples.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 1 milliliter per minute or Null.
StandardMassDetection
Programmatic Pattern: (({RangeP[100*(Gram/Mole), 1200*(Gram/Mole)]..} | All | RangeP[30*(Gram/Mole), 1199*(Gram/Mole), 1*(Gram/Mole)] ;; RangeP[31*(Gram/Mole), 1200*(Gram/Mole), 1*(Gram/Mole)]) | Automatic) | Null
StandardScanTime
The duration of time allowed to pass between each spectral acquisition for Standard sample measurement. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired during the measurement.
Pattern Description: Greater than or equal to 0.05 seconds and less than or equal to 1 second or Null.
StandardProbeTemperature
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius in increments of 1 degree Celsius or Null.
StandardESICapillaryVoltage
The applied voltage differential between the injector and the inlet plate for the mass spectrometry in order to ionize analyte molecules.
Pattern Description: Greater than or equal to 0.3 kilovolts and less than or equal to 1.5 kilovolts or Null.
StandardSamplingConeVoltage
The voltage differential between ion spray entry and the quadrupole mass analyzer for Standard sample measurement.
Programmatic Pattern: ((RangeP[0.1*Volt, 100*Volt] | RangeP[0.1*Volt, 100*Volt] ;; RangeP[0.1*Volt, 100*Volt]) | Automatic) | Null
StandardMassDetectionGain
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
StandardAbsorbanceWavelength
The physical properties of light passed through the flow for the PhotoDiodeArray Detector for Standard measurement.
Programmatic Pattern: ((RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] ;; RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer]) | Automatic) | Null
StandardWavelengthResolution
The increment of wavelength for the range of light passed through the flow for absorbance measurement with the PDA detector for Standard measurement.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
StandardUVFilter
Indicates whether or not to block UV wavelengths (less than 210 nm) from being transmitted through the sample for the PDA detector for Standard measurement.
StandardAbsorbanceSamplingRate
Indicates the frequency of Standard measurement. Lower values will be less susceptible to noise but will record less frequently across time.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
StandardStorageCondition
The non-default conditions under which any standards used by this experiment should be stored after the protocol is completed. If left unset, the standard samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Blanks
Blank
A sample containing just buffer and no analytes, often used to check for background compounds present in the buffer or coming off the column.
Default Calculation: Automatically set from SeparationMode when any other Blank option is specified, otherwise set to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BlankInjectionVolume
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 50 microliters or Null.
BlankFrequency
Specify the frequency at which Blank measurements will be inserted in the measurement sequence. If specified to a number, indicates how often the Blank measurements will run among the analyte samples; for example, if 5 input samples are measured and BlankFrequency is 2, then Blank measurement will occur after the first two samples and again after the third and fourth.
Pattern Description: Greater than 0 in increments of 1 or None, First, Last, FirstAndLast, or GradientChange or Null.
Programmatic Pattern: (((None | First | Last | FirstAndLast | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
BlankColumn
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or list of one or more an object of type or subtype Model[Item, Column] or Object[Item, Column] entries or Null.
Programmatic Pattern: (({ObjectP[{Model[Item, Column], Object[Item, Column]}]..} | ObjectP[{Model[Item, Column], Object[Item, Column]}]) | Automatic) | Null
BlankColumnTemperature
Default Calculation: Automatically set from ColumnTemperature. Automatic resolution can be inherited from the BlankGradient option.
Pattern Description: Ambient or greater than or equal to 20 degrees Celsius and less than or equal to 90 degrees Celsius in increments of 1 degree Celsius or Null.
BlankCO2Gradient
The composition of the supercritical CO2 within the flow, defined for specific time points for Blank samples.
Default Calculation: Automatically set from BlankGradient option or implicitly set from BlankGradientA, BlankGradientB, BlankGradientC, and BlankGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, CO2 Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientA
The composition of CosolventA within the flow, defined for specific time points for Blank measurement.
Default Calculation: Automatically set from BlankGradient option or implicitly set from BlankCO2Gradient, BlankGradientB, BlankGradientC, and BlankGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientB
The composition of CosolventB within the flow, defined for specific time points for Blank measurement.
Default Calculation: Automatically set from BlankGradient option or implicitly set from BlankCO2Gradient, BlankGradientA, BlankGradientC, and BlankGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientC
The composition of CosolventC within the flow, defined for specific time points for Blank measurement.
Default Calculation: Automatically set from BlankGradient option or implicitly set from BlankCO2Gradient, BlankGradientA, BlankGradientB, and BlankGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientD
The composition of CosolventD within the flow, defined for specific time points for Blank measurement.
Default Calculation: Automatically set from BlankGradient option or implicitly set from BlankCO2Gradient, BlankGradientA, BlankGradientB, and BlankGradientC options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankFlowRate
Default Calculation: Automatically set from SeparationMode and Scale or inherited from the method given in the BlankGradient option.
Pattern Description: Greater than or equal to 0 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
BlankBackPressure
The pressure differential between the outlet of the system and the atmosphere for Blank sample measurement.
BlankBackPressure can be changed at different timepoints during the measurement; however, unlike BlankCO2Gradient and BlankFlowRate, BlankBackPressure is not linearly interpolated and, rather, changes stepwise.
Default Calculation: Automatically set to specification within Gradient option; otherwise, set to the same as the first entry in BackPressure.
Pattern Description: Greater than or equal to 1000 pounds‐force per inch squared and less than or equal to 6000 pounds‐force per inch squared or list of one or more {Time, BackPressure} entries or Null.
Programmatic Pattern: ((RangeP[1000*PSI, 6000*PSI] | {{GreaterEqualP[0*Minute], RangeP[1000*PSI, 6000*PSI]}..}) | Automatic) | Null
BlankGradient
The composition over time in the fluid flow for Blank samples. Specific parameters of an object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the BlankGradient options (e.g. BlankCO2Gradient, BlankGradientA, BlankGradientB, BlankGradientC, BlankGradientD, BlankFlowRate, BlankGradientDuration, BlankBackPressure).
Pattern Description: An object of type or subtype Object[Method, SupercriticalFluidGradient] or list of one or more {Time, CO2 Composition, Cosolvent A Composition, Cosolvent B Composition, Cosolvent C Composition, Cosolvent D Composition, Back Pressure, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, SupercriticalFluidGradient]] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], GreaterEqualP[0*PSI], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
BlankGradientDuration
BlankIonMode
BlankMakeupFlowRate
The speed of the MakeupSolvent to supplement column effluent for the mass spectrometry measurement of Blank samples.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 1 milliliter per minute or Null.
BlankMassDetection
Programmatic Pattern: (({RangeP[100*(Gram/Mole), 1200*(Gram/Mole)]..} | All | RangeP[30*(Gram/Mole), 1199*(Gram/Mole), 1*(Gram/Mole)] ;; RangeP[31*(Gram/Mole), 1200*(Gram/Mole), 1*(Gram/Mole)]) | Automatic) | Null
BlankScanTime
The duration of time allowed to pass between each spectral acquisition for Blank sample measurement. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired during the measurement.
Pattern Description: Greater than or equal to 0.05 seconds and less than or equal to 1 second or Null.
BlankProbeTemperature
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius in increments of 1 degree Celsius or Null.
BlankESICapillaryVoltage
The applied voltage differential between the injector and the inlet plate for the mass spectrometry in order to ionize analyte molecules.
Pattern Description: Greater than or equal to 0.3 kilovolts and less than or equal to 1.5 kilovolts or Null.
BlankSamplingConeVoltage
The voltage differential between ion spray entry and the quadrupole mass analyzer for Blank sample measurement.
Programmatic Pattern: ((RangeP[0.1*Volt, 100*Volt] | RangeP[0.1*Volt, 100*Volt] ;; RangeP[0.1*Volt, 100*Volt]) | Automatic) | Null
BlankMassDetectionGain
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
BlankAbsorbanceWavelength
The physical properties of light passed through the flow for the PhotoDiodeArray Detector for Blank measurement.
Programmatic Pattern: ((RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] ;; RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer]) | Automatic) | Null
BlankWavelengthResolution
The increment of wavelength for the range of light passed through the flow for absorbance measurement with the PDA detector for Blank measurement.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
BlankUVFilter
Indicates whether or not to block UV wavelengths (less than 210 nm) from being transmitted through the sample for the PDA detector for Blank measurement.
BlankAbsorbanceSamplingRate
Indicates the frequency of Blank measurement. Lower values will be less susceptible to noise but will record less frequently across time.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
BlankStorageCondition
The non-default conditions under which any Blank samples used by this experiment should be stored after the protocol is completed. If left unset, the standard samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
ColumnPrime
ColumnRefreshFrequency
Specify the frequency at which procedures to clear out and re-prime the column will be inserted into the order of analyte injections. If specified to a number, indicates how often the column prime runs will run among the analyte samples; for example, if 5 input samples are measured and ColumnRefreshFrequency is 2, then a column prime will occur after the first two samples and again after the third and fourth.
Default Calculation: Set to Null when InjectionTable option is specified (meaning that this option is inconsequential); otherwise, set to FirstAndLast (meaning initial column prime before the measurements and final column flush after measurements.) when there is a Column. Set to None if there is no Column (meaning no column flush).
ColumnPrimeColumnTemperature
Default Calculation: Automatically set from ColumnTemperature. Automatic resolution can be inherited from the ColumnPrimeGradient option.
Pattern Description: Ambient or greater than or equal to 20 degrees Celsius and less than or equal to 90 degrees Celsius in increments of 1 degree Celsius or Null.
ColumnPrimeCO2Gradient
The composition of the supercritical CO2 within the flow, defined for specific time points for ColumnPrime runs.
Default Calculation: Automatically set from ColumnPrimeGradient option or implicitly set from ColumnPrimeGradientA, ColumnPrimeGradientB, ColumnPrimeGradientC, and ColumnPrimeGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, CO2 Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeGradientA
The composition of CosolventA within the flow, defined for specific time points for ColumnPrime runs.
Default Calculation: Automatically set from ColumnPrimeGradient option or implicitly set from ColumnPrimeCO2Gradient, ColumnPrimeGradientB, ColumnPrimeGradientC, and ColumnPrimeGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeGradientB
The composition of CosolventB within the flow, defined for specific time points for ColumnPrime runs.
Default Calculation: Automatically set from ColumnPrimeGradient option or implicitly set from ColumnPrimeCO2Gradient, ColumnPrimeGradientA, ColumnPrimeGradientC, and ColumnPrimeGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeGradientC
The composition of CosolventC within the flow, defined for specific time points for ColumnPrime runs.
Default Calculation: Automatically set from ColumnPrimeGradient option or implicitly set from ColumnPrimeCO2Gradient, ColumnPrimeGradientA, ColumnPrimeGradientB, and ColumnPrimeGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeGradientD
The composition of CosolventD within the flow, defined for specific time points for ColumnPrime runs.
Default Calculation: Automatically set from ColumnPrimeGradient option or implicitly set from ColumnPrimeCO2Gradient, ColumnPrimeGradientA, ColumnPrimeGradientB, and ColumnPrimeGradientC options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeFlowRate
Default Calculation: Automatically set from SeparationMode and Scale or inherited from the method given in the ColumnPrimeGradient option.
Pattern Description: Greater than or equal to 0 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnPrimeGradientDuration
ColumnPrimeGradient
The composition over time in the fluid flow for ColumnPrime runs. Specific parameters of an object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the ColumnPrimeGradient options (e.g. ColumnPrimeCO2Gradient, ColumnPrimeGradientA, ColumnPrimeGradientB, ColumnPrimeGradientC, ColumnPrimeGradientD, ColumnPrimeFlowRate, ColumnPrimeGradientDuration, ColumnPrimeBackPressure).
Pattern Description: An object of type or subtype Object[Method, SupercriticalFluidGradient] or list of one or more {Time, CO2 Composition, Cosolvent A Composition, Cosolvent B Composition, Cosolvent C Composition, Cosolvent D Composition, Back Pressure, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, SupercriticalFluidGradient]] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], GreaterEqualP[0*PSI], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnPrimeBackPressure
The pressure differential between the outlet of the system and the atmosphere for ColumnPrime runs.
ColumnPrimeBackPressure can be changed at different timepoints during the measurement; however, unlike ColumnPrimeCO2Gradient and ColumnPrimeFlowRate, ColumnPrimeBackPressure is not linearly interpolated and, rather, changes stepwise.
Default Calculation: Automatically set to specification within Gradient option; otherwise, set to the same as the first entry in BackPressure.
Pattern Description: Greater than or equal to 1000 pounds‐force per inch squared and less than or equal to 6000 pounds‐force per inch squared or list of one or more {Time, BackPressure} entries or Null.
Programmatic Pattern: ((RangeP[1000*PSI, 6000*PSI] | {{GreaterEqualP[0*Minute], RangeP[1000*PSI, 6000*PSI]}..}) | Automatic) | Null
ColumnPrimeIonMode
ColumnPrimeMakeupFlowRate
The speed of the MakeupSolvent to supplement column effluent for the mass spectrometry measurement of ColumnPrime runs.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 1 milliliter per minute or Null.
ColumnPrimeMassDetection
Programmatic Pattern: (({RangeP[100*(Gram/Mole), 1200*(Gram/Mole)]..} | All | RangeP[30*(Gram/Mole), 1199*(Gram/Mole), 1*(Gram/Mole)] ;; RangeP[31*(Gram/Mole), 1200*(Gram/Mole), 1*(Gram/Mole)]) | Automatic) | Null
ColumnPrimeScanTime
The duration of time allowed to pass between each spectral acquisition for ColumnPrime runs. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired during the measurement.
Pattern Description: Greater than or equal to 0.05 seconds and less than or equal to 1 second or Null.
ColumnPrimeProbeTemperature
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius in increments of 1 degree Celsius or Null.
ColumnPrimeESICapillaryVoltage
The applied voltage differential between the injector and the inlet plate for the mass spectrometry in order to ionize analyte molecules for each column prime.
Pattern Description: Greater than or equal to 0.3 kilovolts and less than or equal to 1.5 kilovolts or Null.
ColumnPrimeSamplingConeVoltage
The voltage differential between ion spray entry and the quadrupole mass analyzer for ColumnPrime runs.
Programmatic Pattern: ((RangeP[0.1*Volt, 100*Volt] | RangeP[0.1*Volt, 100*Volt] ;; RangeP[0.1*Volt, 100*Volt]) | Automatic) | Null
ColumnPrimeMassDetectionGain
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
ColumnPrimeAbsorbanceWavelength
The physical properties of light passed through the flow for the PhotoDiodeArray Detector for ColumnPrime runs.
Programmatic Pattern: ((RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] ;; RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer]) | Automatic) | Null
ColumnPrimeWavelengthResolution
The increment of wavelength for the range of light passed through the flow for absorbance measurement with the PDA detector for ColumnPrime runs.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
ColumnPrimeUVFilter
Indicates whether or not to block UV wavelengths (less than 210 nm) from being transmitted through the sample for the PDA detector for ColumnPrime runs.
ColumnPrimeAbsorbanceSamplingRate
Indicates the frequency of ColumnPrime runs. Lower values will be less susceptible to noise but will record less frequently across time.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
ColumnFlush
ColumnFlushColumnTemperature
Default Calculation: Automatically set from ColumnTemperature. Automatic resolution can be inherited from the ColumnFlushGradient option.
Pattern Description: Ambient or greater than or equal to 20 degrees Celsius and less than or equal to 90 degrees Celsius in increments of 1 degree Celsius or Null.
ColumnFlushCO2Gradient
The composition of the supercritical CO2 within the flow, defined for specific time points for ColumnFlush runs.
Default Calculation: Automatically set from ColumnFlushGradient option or implicitly set from ColumnFlushGradientA, ColumnFlushGradientB, ColumnFlushGradientC, and ColumnFlushGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, CO2 Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushGradientA
The composition of CosolventA within the flow, defined for specific time points for ColumnFlush runs.
Default Calculation: Automatically set from ColumnFlushGradient option or implicitly set from ColumnFlushCO2Gradient, ColumnFlushGradientB, ColumnFlushGradientC, and ColumnFlushGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushGradientB
The composition of CosolventB within the flow, defined for specific time points for ColumnFlush runs.
Default Calculation: Automatically set from ColumnFlushGradient option or implicitly set from ColumnFlushCO2Gradient, ColumnFlushGradientA, ColumnFlushGradientC, and ColumnFlushGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushGradientC
The composition of CosolventC within the flow, defined for specific time points for ColumnFlush runs.
Default Calculation: Automatically set from ColumnFlushGradient option or implicitly set from ColumnFlushCO2Gradient, ColumnFlushGradientA, ColumnFlushGradientB, and ColumnFlushGradientD options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushGradientD
The composition of CosolventD within the flow, defined for specific time points for ColumnFlush runs.
Default Calculation: Automatically set from ColumnFlushGradient option or implicitly set from ColumnFlushCO2Gradient, ColumnFlushGradientA, ColumnFlushGradientB, and ColumnFlushGradientC options.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Cosolvent D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushFlowRate
Default Calculation: Automatically set from SeparationMode and Scale or inherited from the method given in the ColumnFlushGradient option.
Pattern Description: Greater than or equal to 0 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(Milliliter/Minute)] | {{GreaterEqualP[0*Minute], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnFlushGradientDuration
ColumnFlushGradient
The composition over time in the fluid flow for ColumnFlush runs. Specific parameters of an object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the ColumnFlushGradient options (e.g. ColumnFlushCO2Gradient, ColumnFlushGradientA, ColumnFlushGradientB, ColumnFlushGradientC, ColumnFlushGradientD, ColumnFlushFlowRate, ColumnFlushGradientDuration, ColumnFlushBackPressure).
Pattern Description: An object of type or subtype Object[Method, SupercriticalFluidGradient] or list of one or more {Time, CO2 Composition, Cosolvent A Composition, Cosolvent B Composition, Cosolvent C Composition, Cosolvent D Composition, Back Pressure, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, SupercriticalFluidGradient]] | {{GreaterEqualP[0*Minute], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], GreaterEqualP[0*PSI], GreaterEqualP[0*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnFlushBackPressure
The pressure differential between the outlet of the system and the atmosphere for ColumnFlush runs.
ColumnFlushBackPressure can be changed at different timepoints during the measurement; however, unlike ColumnFlushCO2Gradient and ColumnFlushFlowRate, ColumnFlushBackPressure is not linearly interpolated and, rather, changes stepwise.
Default Calculation: Automatically set to specification within Gradient option; otherwise, set to the same as the first entry in BackPressure.
Pattern Description: Greater than or equal to 1000 pounds‐force per inch squared and less than or equal to 6000 pounds‐force per inch squared or list of one or more {Time, BackPressure} entries or Null.
Programmatic Pattern: ((RangeP[1000*PSI, 6000*PSI] | {{GreaterEqualP[0*Minute], RangeP[1000*PSI, 6000*PSI]}..}) | Automatic) | Null
ColumnFlushIonMode
ColumnFlushMakeupFlowRate
The speed of the MakeupSolvent to supplement column effluent for the mass spectrometry measurement of ColumnFlush runs.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 1 milliliter per minute or Null.
ColumnFlushMassDetection
Programmatic Pattern: (({RangeP[100*(Gram/Mole), 1200*(Gram/Mole)]..} | All | RangeP[30*(Gram/Mole), 1199*(Gram/Mole), 1*(Gram/Mole)] ;; RangeP[31*(Gram/Mole), 1200*(Gram/Mole), 1*(Gram/Mole)]) | Automatic) | Null
ColumnFlushScanTime
The duration of time allowed to pass between each spectral acquisition for ColumnFlush runs. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired during the measurement.
Pattern Description: Greater than or equal to 0.05 seconds and less than or equal to 1 second or Null.
ColumnFlushProbeTemperature
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius in increments of 1 degree Celsius or Null.
ColumnFlushESICapillaryVoltage
The applied voltage differential between the injector and the inlet plate for the mass spectrometry in order to ionize analyte molecules for each column flush.
Pattern Description: Greater than or equal to 0.3 kilovolts and less than or equal to 1.5 kilovolts or Null.
ColumnFlushSamplingConeVoltage
The voltage differential between ion spray entry and the quadrupole mass analyzer for ColumnFlush runs.
Programmatic Pattern: ((RangeP[0.1*Volt, 100*Volt] | RangeP[0.1*Volt, 100*Volt] ;; RangeP[0.1*Volt, 100*Volt]) | Automatic) | Null
ColumnFlushMassDetectionGain
Pattern Description: Greater than or equal to 1 and less than or equal to 10 in increments of 1 or Null.
ColumnFlushAbsorbanceWavelength
The physical properties of light passed through the flow for the PhotoDiodeArray Detector for ColumnFlush runs.
Programmatic Pattern: ((RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer] ;; RangeP[190*Nanometer, 800*Nanometer, 1*Nanometer]) | Automatic) | Null
ColumnFlushWavelengthResolution
The increment of wavelength for the range of light passed through the flow for absorbance measurement with the PDA detector for ColumnFlush runs.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
ColumnFlushUVFilter
Indicates whether or not to block UV wavelengths (less than 210 nm) from being transmitted through the sample for the PDA detector for ColumnFlush runs.
ColumnFlushAbsorbanceSamplingRate
Indicates the frequency of ColumnFlush runs. Lower values will be less susceptible to noise but will record less frequently across time.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
Model Input
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Programmatic Pattern: ((Null | (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All)) | Automatic) | Null
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise, automatically set to Model[Container, Plate, "96-well 2mL Deep Well Plate"].
Post Experiment
SamplesInStorageCondition
The non-default conditions under which the SamplesIn of this experiment should be stored after the protocol is completed. If left unset, SamplesIn will be stored according to their current StorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Sample Prep Options
Sample Preparation
PreparatoryUnitOperations
Specifies a sequence of transferring, aliquoting, consolidating, or mixing of new or existing samples before the main experiment. These prepared samples can be used in the main experiment by referencing their defined name. For more information, please reference the documentation for ExperimentSamplePreparation.
Pattern Description: List of one or more unit Operation ManualSamplePreparation or RoboticSamplePreparation or unit Operation must match SamplePreparationP entries or Null.
Programmatic Pattern: {((ManualSamplePreparationMethodP | RoboticSamplePreparationMethodP) | SamplePreparationP)..} | Null
Preparatory Incubation
Incubate
Indicates if the SamplesIn should be incubated at a fixed temperature prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Incubation options are set. Otherwise, resolves to False.
IncubationTemperature
Temperature at which the SamplesIn should be incubated for the duration of the IncubationTime prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -20 degrees Celsius and less than or equal to 500 degrees Celsius or Null.
Programmatic Pattern: ((Ambient | RangeP[$MinIncubationTemperature, $MaxIncubationTemperature]) | Automatic) | Null
IncubationTime
Duration for which SamplesIn should be incubated at the IncubationTemperature, prior to starting the experiment.
Mix
Default Calculation: Automatically resolves to True if any Mix related options are set. Otherwise, resolves to False.
MixType
Default Calculation: Automatically resolves based on the container of the sample and the Mix option.
Pattern Description: Roll, Vortex, Sonicate, Pipette, Invert, Stir, Shake, Homogenize, Swirl, Disrupt, or Nutate or Null.
MixUntilDissolved
Indicates if the mix should be continued up to the MaxIncubationTime or MaxNumberOfMixes (chosen according to the mix Type), in an attempt dissolve any solute. Any mixing/incubation will occur prior to starting the experiment.
Default Calculation: Automatically resolves to True if MaxIncubationTime or MaxNumberOfMixes is set.
MaxIncubationTime
Maximum duration of time for which the samples will be mixed while incubated in an attempt to dissolve any solute, if the MixUntilDissolved option is chosen. This occurs prior to starting the experiment.
Default Calculation: Automatically resolves based on MixType, MixUntilDissolved, and the container of the given sample.
IncubationInstrument
Default Calculation: Automatically resolves based on the options Mix, Temperature, MixType and container of the sample.
Pattern Description: An object of type or subtype Model[Instrument, Roller], Model[Instrument, OverheadStirrer], Model[Instrument, Vortex], Model[Instrument, Shaker], Model[Instrument, BottleRoller], Model[Instrument, Roller], Model[Instrument, Sonicator], Model[Instrument, HeatBlock], Model[Instrument, Homogenizer], Model[Instrument, Disruptor], Model[Instrument, Nutator], Model[Instrument, Thermocycler], Model[Instrument, EnvironmentalChamber], Model[Instrument, Pipette], Object[Instrument, Roller], Object[Instrument, OverheadStirrer], Object[Instrument, Vortex], Object[Instrument, Shaker], Object[Instrument, BottleRoller], Object[Instrument, Roller], Object[Instrument, Sonicator], Object[Instrument, HeatBlock], Object[Instrument, Homogenizer], Object[Instrument, Disruptor], Object[Instrument, Nutator], Object[Instrument, Thermocycler], Object[Instrument, EnvironmentalChamber], or Object[Instrument, Pipette] or Null.
AnnealingTime
Minimum duration for which the SamplesIn should remain in the incubator allowing the system to settle to room temperature after the IncubationTime has passed but prior to starting the experiment.
IncubateAliquotContainer
The desired type of container that should be used to prepare and house the incubation samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
IncubateAliquotDestinationWell
The desired position in the corresponding IncubateAliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
IncubateAliquot
The amount of each sample that should be transferred from the SamplesIn into the IncubateAliquotContainer when performing an aliquot before incubation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Centrifugation
Centrifuge
Indicates if the SamplesIn should be centrifuged prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Centrifuge options are set. Otherwise, resolves to False.
CentrifugeInstrument
Pattern Description: An object of type or subtype Model[Instrument, Centrifuge] or Object[Instrument, Centrifuge] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, Centrifuge], Object[Instrument, Centrifuge]}] | Automatic) | Null
CentrifugeIntensity
The rotational speed or the force that will be applied to the samples by centrifugation prior to starting the experiment.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
CentrifugeTime
CentrifugeTemperature
The temperature at which the centrifuge chamber should be held while the samples are being centrifuged prior to starting the experiment.
Pattern Description: Ambient or greater than or equal to -10 degrees Celsius and less than or equal to 40 degrees Celsius or Null.
CentrifugeAliquotContainer
The desired type of container that should be used to prepare and house the centrifuge samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
CentrifugeAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
CentrifugeAliquot
The amount of each sample that should be transferred from the SamplesIn into the CentrifugeAliquotContainer when performing an aliquot before centrifugation.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
Preparatory Filtering
Filtration
Indicates if the SamplesIn should be filter prior to starting the experiment or any aliquoting. Sample Preparation occurs in the order of Incubation, Centrifugation, Filtration, and then Aliquoting (if specified).
Default Calculation: Resolves to True if any of the corresponding Filter options are set. Otherwise, resolves to False.
FiltrationType
Default Calculation: Will automatically resolve to a filtration type appropriate for the volume of sample being filtered.
FilterInstrument
Default Calculation: Will automatically resolved to an instrument appropriate for the filtration type.
Pattern Description: An object of type or subtype Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], or Object[Instrument, SyringePump] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterBlock], Object[Instrument, FilterBlock], Model[Instrument, PeristalticPump], Object[Instrument, PeristalticPump], Model[Instrument, VacuumPump], Object[Instrument, VacuumPump], Model[Instrument, Centrifuge], Object[Instrument, Centrifuge], Model[Instrument, SyringePump], Object[Instrument, SyringePump]}] | Automatic) | Null
Filter
The filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Will automatically resolve to a filter appropriate for the filtration type and instrument.
Pattern Description: An object of type or subtype Model[Container, Plate, Filter], Model[Container, Vessel, Filter], or Model[Item, Filter] or Null.
Programmatic Pattern: (ObjectP[{Model[Container, Plate, Filter], Model[Container, Vessel, Filter], Model[Item, Filter]}] | Automatic) | Null
FilterMaterial
The membrane material of the filter that should be used to remove impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter material for the given sample is Filtration is set to True.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
PrefilterMaterial
The material from which the prefilter filtration membrane should be made of to remove impurities from the SamplesIn prior to starting the experiment.
Pattern Description: Cellulose, Cotton, Polyethylene, Polypropylene, PTFE, Nylon, PES, PLUS, PVDF, GlassFiber, GHP, UHMWPE, EPDM, DuraporePVDF, GxF, ZebaDesaltingResin, NickelResin, AgaroseResin, CobaltResin, Silica, HLB, or AnoporeAlumina or Null.
FilterPoreSize
The pore size of the filter that should be used when removing impurities from the SamplesIn prior to starting the experiment.
Default Calculation: Resolves to an appropriate filter pore size for the given sample is Filtration is set to True.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
PrefilterPoreSize
The pore size of the filter; all particles larger than this should be removed during the filtration.
Pattern Description: 0.008 micrometers, 0.02 micrometers, 0.1 micrometers, 0.2 micrometers, 0.22 micrometers, 0.45 micrometers, 1. micrometer, 1.1 micrometers, 2.5 micrometers, 6. micrometers, 20. micrometers, 30. micrometers, or 100. micrometers or Null.
FilterSyringe
Default Calculation: Resolves to an syringe appropriate to the volume of sample being filtered, if Filtration is set to True.
Pattern Description: An object of type or subtype Model[Container, Syringe] or Object[Container, Syringe] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Container, Syringe], Object[Container, Syringe]}] | _String) | Automatic) | Null
FilterHousing
The filter housing that should be used to hold the filter membrane when filtration is performed using a standalone filter membrane.
Default Calculation: Resolve to an housing capable of holding the size of the membrane being used, if filter with Membrane FilterType is being used and Filtration is set to True.
Pattern Description: An object of type or subtype Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], or Object[Instrument, FilterBlock] or Null.
Programmatic Pattern: (ObjectP[{Model[Instrument, FilterHousing], Object[Instrument, FilterHousing], Model[Instrument, FilterBlock], Object[Instrument, FilterBlock]}] | Automatic) | Null
FilterIntensity
Default Calculation: Will automatically resolve to 2000 GravitationalAcceleration if FiltrationType is Centrifuge and Filtration is True.
Pattern Description: Greater than 0 revolutions per minute or greater than 0 standard accelerations due to gravity on the surface of the earth or Null.
Programmatic Pattern: ((GreaterP[0*RPM] | GreaterP[0*GravitationalAcceleration]) | Automatic) | Null
FilterTime
Default Calculation: Will automatically resolve to 5 Minute if FiltrationType is Centrifuge and Filtration is True.
FilterTemperature
The temperature at which the centrifuge chamber will be held while the samples are being centrifuged during filtration.
Default Calculation: Will automatically resolve to 22 Celsius if FiltrationType is Centrifuge and Filtration is True.
FilterContainerOut
The desired container filtered samples should be produced in or transferred into by the end of filtration, with indices indicating grouping of samples in the same plates, if desired.
Default Calculation: Automatically set as the PreferredContainer for the Volume of the sample. For plates, attempts to fill all wells of a single plate with the same model before using another one.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or {Index, Container} or Null.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | {GreaterEqualP[1, 1] | Automatic, (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquotDestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
FilterAliquotContainer
The desired type of container that should be used to prepare and house the filter samples which should be used in lieu of the SamplesIn for the experiment.
Programmatic Pattern: ((ObjectP[Model[Container]] | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | Automatic}) | Automatic) | Null
FilterAliquot
The amount of each sample that should be transferred from the SamplesIn into the FilterAliquotContainer when performing an aliquot before filtration.
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container.
Pattern Description: All or greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
FilterSterile
Default Calculation: Resolve to False if Filtration is indicated. If sterile filtration is desired, this option must manually be set to True.
Aliquoting
Aliquot
Indicates if aliquots should be taken from the SamplesIn and transferred into new AliquotSamples used in lieu of the SamplesIn for the experiment. Note that if NumberOfReplicates is specified this indicates that the input samples will also be aliquoted that number of times. Note that Aliquoting (if specified) occurs after any Sample Preparation (if specified).
AliquotAmount
Default Calculation: Automatically set as the smaller between the current sample volume and the maximum volume of the destination container if a liquid, or the current Mass or Count if a solid or counted item, respectively.
Programmatic Pattern: ((RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All) | Automatic) | Null
TargetConcentration
The desired final concentration of analyte in the AliquotSamples after dilution of aliquots of SamplesIn with the ConcentratedBuffer and BufferDiluent which should be used in lieu of the SamplesIn for the experiment.
TargetConcentrationAnalyte
Default Calculation: Automatically set to the first value in the Analytes field of the input sample, or, if not populated, to the first analyte in the Composition field of the input sample, or if none exist, the first identity model of any kind in the Composition field.
Pattern Description: An object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] or Null.
AssayVolume
Default Calculation: Automatically determined based on Volume and TargetConcentration option values.
Pattern Description: Greater than or equal to 1 microliter and less than or equal to 20 liters or Null.
ConcentratedBuffer
The concentrated buffer which should be diluted by the BufferDilutionFactor in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
BufferDilutionFactor
The dilution factor by which the concentrated buffer should be diluted in the final solution (i.e., the combination of the sample, ConcentratedBuffer, and BufferDiluent). The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: If ConcentratedBuffer is specified, automatically set to the ConcentratedBufferDilutionFactor of that sample; otherwise, set to Null.
BufferDiluent
The buffer used to dilute the aliquot sample such that ConcentratedBuffer is diluted by BufferDilutionFactor in the final solution. The ConcentratedBuffer and BufferDiluent will be combined and then mixed with the sample, where the combined volume of these buffers is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AssayBuffer
The buffer that should be added to any aliquots requiring dilution, where the volume of this buffer added is the difference between the AliquotAmount and the total AssayVolume.
Default Calculation: Automatically resolves to Model[Sample, "Milli-Q water"] if ConcentratedBuffer is not specified; otherwise, resolves to Null.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
AliquotSampleStorageCondition
The non-default conditions under which any aliquot samples generated by this experiment should be stored after the protocol is completed.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
DestinationWell
The desired position in the corresponding AliquotContainer in which the aliquot samples will be placed.
Default Calculation: Automatically resolves to A1 in containers with only one position. For plates, fills wells in the order provided by the function AllWells.
Pattern Description: Any well from A1 to H12 or list of one or more any well from A1 to H12 or any well from A1 to H12 entries or Null.
Programmatic Pattern: ((WellPositionP | {((Automatic | Null) | WellPositionP)..}) | Automatic) | Null
AliquotContainer
The desired type of container that should be used to prepare and house the aliquot samples, with indices indicating grouping of samples in the same plates, if desired. This option will resolve to be the length of the SamplesIn * NumberOfReplicates.
Default Calculation: Automatically set as the PreferredContainer for the AssayVolume of the sample. For plates, attempts to fill all wells of a single plate with the same model before aliquoting into the next.
Pattern Description: An object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null or {Index, Container} or list of one or more an object of type or subtype Model[Container] or Object[Container] or a prepared sample or Automatic or Null entries or list of one or more Automatic or Null or {Index, Container} entries.
Programmatic Pattern: (((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null) | {GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | {((ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null))..} | {({GreaterEqualP[1, 1] | (Automatic | Null), (ObjectP[{Model[Container], Object[Container]}] | _String) | (Automatic | Null)} | (Automatic | Null))..}) | Automatic) | Null
AliquotPreparation
Default Calculation: Automatic resolution will occur based on manipulation volumes and container types.
ConsolidateAliquots
Protocol Options
Organizational Information
Template
A template protocol whose methodology should be reproduced in running this experiment. Option values will be inherited from the template protocol, but can be individually overridden by directly specifying values for those options to this Experiment function.
Pattern Description: An object of type or subtype Object[Protocol] or an object of type or subtype of Object[Protocol] with UnresolvedOptions, ResolvedOptions specified or Null.
Programmatic Pattern: (ObjectP[Object[Protocol]] | FieldReferenceP[Object[Protocol], {UnresolvedOptions, ResolvedOptions}]) | Null
Name
A object name which should be used to refer to the output object in lieu of an automatically generated ID number.
Post Experiment
MeasureWeight
Indicates if any solid samples that are modified in the course of the experiment should have their weights measured and updated after running the experiment. Please note that public samples are weighed regardless of the value of this option.
MeasureVolume
Indicates if any liquid samples that are modified in the course of the experiment should have their volumes measured and updated after running the experiment. Please note that public samples are volume measured regardless of the value of this option.
ImageSample
Example Calls
Basics
Measurement involves supercritical carbon dioxide as the primary mobile phase, perfect for analytes with poor solubility in water. To run a SupercriticalFluidChromatography experiment, simply run:
Separation
Cosolvents can also be added to the fluid stream in order to change solubility and column retention properties. For example, methanol can be added in gradient fashion to the mobile phase. In the following experiment, the mobile phase changes from 95% carbon dioxide/5% methanol to 60% carbon dioxide/40% methanol by volume over the course of 5 minutes for each sample injection:
Column Selection
Multiple columns can be installed into the instrument and used individually for different sample injections, enabling rapid scouting of the ideal chromatographic conditions:
Standards and Blanks
ExperimentSupercriticalFluidChromatography can be used to quantify analytes in a sample, in which case, a Standard sample is employed to serve as reference. A standard can be submitted before and after the injection sequence of the samples simply by running:
Likewise, a Blank sample can be used to see if there is any background from the injection process. To run a blank to occur between every 5 samples, use the following command:
Detection
The absorbance and mass spectrometer measurement can be controlled as well. For example, a range of wavelengths and specific masses can be requested:
Injection Table
Data Processing
Warnings and Errors
Messages (30)
BackPressurePrecision (1)
ColumnSelectorConflict (1)
CosolventConflict (1)
FlowRatePrecision (1)
GradientPercentPrecision (1)
GradientShortcutAmbiguity (1)
GradientShortcutConflict (1)
GradientTimePrecision (1)
InjectionTableBlankConflict (1)
InjectionTableBlankFrequencyConflict (1)
InjectionTableColumnConflictSFC (1)
InjectionTableColumnGradientConflict (1)
InjectionTableForeignSamples (1)
InjectionTableGradientConflict (1)
InjectionTableStandardConflict (1)
InjectionTableStandardFrequencyConflict (1)
NonbinaryGradientDefined (1)
ObjectDoesNotExist (4)
SelectorInjectionTableConflict (1)
SharedContainerStorageCondition (1)
WavelengthResolutionAdjusted (2)
Possible Issues
Equilibration
Equilibrate to the correct buffer conditions at the end of each gradient to reach a state that is appropriate for the start of the next gradient.
Column flush
The column will end up being stored in whatever the final gradient solvent conditions are so that should be considered for column protection.
Minimize water
Last modified on Wed 17 Sep 2025 16:26:57