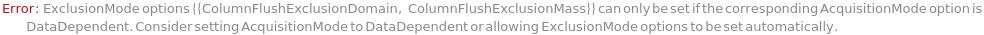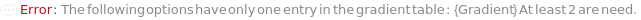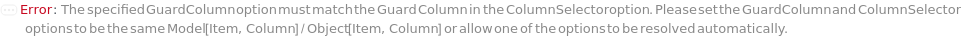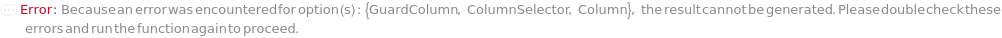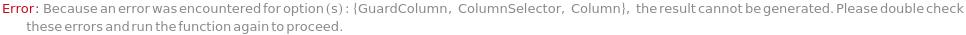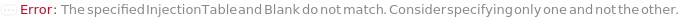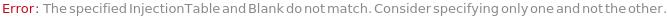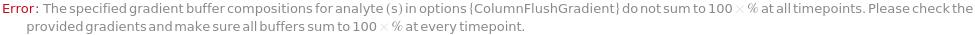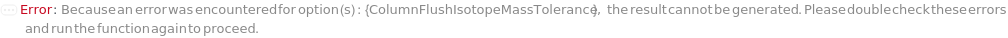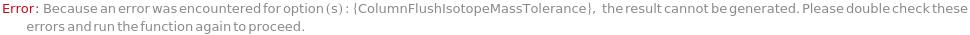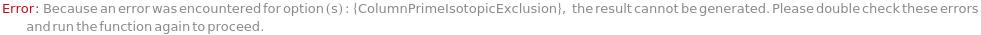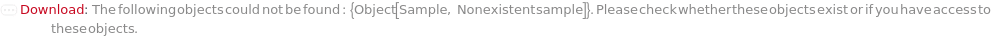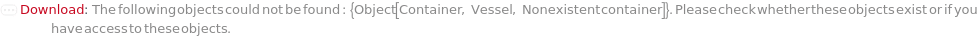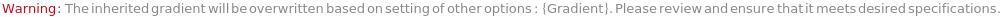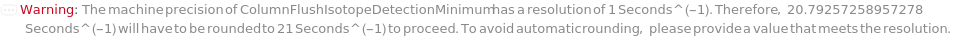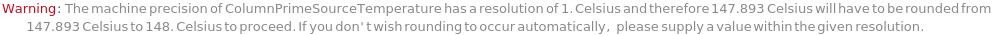General
SeparationMode
The category of method used to elicit differential column retention due to interaction between molecules in the mobile phase with those on the stationary phase (column).
Default Calculation: Automatically set to match the Separation Mode listed with the provided column.
Pattern Description: NormalPhase, ReversePhase, IonExchange, SizeExclusion, Affinity, or Chiral.
Programmatic Pattern: SeparationModeP | Automatic
MassAnalyzer
The manner of detection used to resolve and detect molecules. QTOF accelerates ions through an elongated flight tube and separates ions by their flight time (related to mass to charge ratio).
Default Calculation: Is automatically set to the QTOF.
Pattern Description: QTOF or TripleQuadrupole.
Programmatic Pattern: (QTOF | TripleQuadrupole) | Automatic
ChromatographyInstrument
The device used to separate molecules from the sample mixture using mobile liquid through an adsorbent column.
Default Value: Model[Instrument, HPLC, Waters Acquity UPLC I-Class PDA]
Default Calculation: Automatically set to an instrument model that is available for the best MassSpectrometerInstrument.
Pattern Description: An object of type or subtype Model[Instrument, HPLC] or Object[Instrument, HPLC]
Programmatic Pattern: ObjectP[{Model[Instrument, HPLC], Object[Instrument, HPLC]}]
MassSpectrometerInstrument
The device used to ionize, separate, fragment (optionally), and detect analyte species.
Default Calculation: Is automatically set according to MassAnalyzer: Model[Instrument, MassSpectrometer, "Xevo G2-XS QTOF"] for QTOF, Model[Instrument, MassSpectrometer, "QTRAP 6500"] for using TripleQuadrupole.
Pattern Description: An object of type or subtype Model[Instrument, MassSpectrometer] or Object[Instrument, MassSpectrometer]
Programmatic Pattern: ObjectP[{Model[Instrument, MassSpectrometer], Object[Instrument, MassSpectrometer]}] | Automatic
Detector
Additional measurements to employ in concert with MassSpectrometry. Other measurements include: PhotoDiodeArray (measures the absorbance of a range of wavelengths), system Temperature (QTOF only), and system Pressure (QTOF only).
Default Value: {Temperature, Pressure, PhotoDiodeArray}
Pattern Description: List of one or more Temperature, Pressure, or PhotoDiodeArray entries or Temperature, Pressure, or PhotoDiodeArray.
Programmatic Pattern: (Temperature | Pressure | PhotoDiodeArray) | {(Temperature | Pressure | PhotoDiodeArray)..}
GuardColumn
The protective device placed in the flow path before the Column in order to adsorb fouling contaminants and, thus, preserve the Column lifetime.
Default Calculation: Automatically set from the column model's PreferredGuardColumn. If Column is Null, GuardColumn is automatically set to Null.
Pattern Description: An object of type or subtype Model[Item, Column], Object[Item, Column], Model[Item, Cartridge, Column], or Object[Item, Cartridge, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column], Model[Item, Cartridge, Column], Object[Item, Cartridge, Column]}] | Automatic) | Null
NumberOfReplicates
The number of times to repeat measurements on each provided sample(s). If Aliquot -> True, this also indicates the number of times each provided sample will be aliquoted. For experiment samples {A,B,C} if NumberOfReplicates is specified as 3, the order of samples to run on the instrument will be {A,A,A,B,B,B,C,C,C}.
Pattern Description: Greater than or equal to 1 and less than or equal to 96 in increments of 1 or Null.
Programmatic Pattern: RangeP[1, 96, 1] | Null
Chromatography
Column
The item containing the stationary phase through which the mobile phase and input samples flow. It adsorbs and separates the molecules within the sample based on the properties of the mobile phase, Samples, Column material, and Column Temperature.
Default Calculation: Automatically set to a column model compatible for the instrument selected and specified separation Mode.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic) | Null
SecondaryColumn
The additional stationary phase through which the mobile phase and input samples flow. The SecondaryColumn selectively adsorb analytes and is connected to flow path, downstream of the Column.
Default Calculation: If ColumnSelector is specified, set from there; otherwise, set to Null.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic) | Null
TertiaryColumn
The additional stationary phase through which the mobile phase and input samples flow. The TertiaryColumn selectively adsorb analytes and is connected to flow path, downstream of the SecondaryColumn.
Default Calculation: If ColumnSelector is specified, set from there; otherwise, set to Null.
Pattern Description: An object of type or subtype Model[Item, Column] or Object[Item, Column] or Null.
Programmatic Pattern: (ObjectP[{Model[Item, Column], Object[Item, Column]}] | Automatic) | Null
ColumnSelector
The set of all the columns loaded into the Instrument's column selector and referenced in Column, SecondaryColumn, TertiaryColumn. The Serial configuration indicates one fluid line for all the samples, Standard and Blank. The Selector configuration indicates use of a column selector where the column line is programmatically switchable between samples.
Default Calculation: Automatically set to match the values in Column, SecondaryColumn, TertiaryColumn and GuardColumn options.
Pattern Description: {Guard Column, Column, Secondary Column, Tertiary Column}
Programmatic Pattern: {ObjectP[{Model[Item, Column], Object[Item, Column]}] | (Automatic | Null), ObjectP[{Model[Item, Column], Object[Item, Column]}] | (Automatic | Null), ObjectP[{Model[Item, Column], Object[Item, Column]}] | (Automatic | Null), ObjectP[{Model[Item, Column], Object[Item, Column]}] | (Automatic | Null)} | Automatic
BufferA
A solvent or buffer placed in the 'A' bottle as shown in Figure 2.1.1 of ExperimentLCMS help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientA option.
Default Calculation: Automatically set from SeparationMode (e.g. Water buffer if ReversePhase) or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
BufferB
A solvent or buffer placed in the 'B' bottle as shown in Figure 2.1.1 of ExperimentLCMS help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientB option.
Default Calculation: Automatically set from SeparationMode (e.g. Acetonitrile buffer if ReversePhase) or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
BufferC
A solvent or buffer placed in the 'C' bottle as shown in Figure 2.1.1 of ExperimentLCMS help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientC option.
Default Calculation: Automatically set from SeparationMode or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
BufferD
A solvent or buffer placed in the 'D' bottle as shown in Figure 2.1.1 of ExperimentLCMS help file, pumped through the instrument as part of the mobile phase, the compositions of which is determined by the GradientD option.
Default Calculation: Automatically set from SeparationMode or the objects specified by the Gradient option.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
Sample Parameters
InjectionTable
The order of Sample, Standard and Blank sample injected into the Instrument during measurement and/or collection. This also includes the priming and flushing of the column(s).
Default Calculation: Samples are inserted in the order of the input samples based with the number of replicates. Standard and Blank samples are inserted based on the determination of StandardFrequency and BlankFrequency options. For example, StandardFrequency -> FirstAndLast and BlankFrequency -> Null result in Standard samples injected first, then samples, and then the Standard sample set again at the end. Column priming is inserted at the beginning and repeated according to ColumnPrimeFrequency. Column flushing is inserted at the end.
Pattern Description: List of one or more {Type, Sample, InjectionVolume, Column Temperature, Gradient, Mass Spectrometry} entries.
Programmatic Pattern: {{Standard | Sample | Blank | ColumnPrime | ColumnFlush, (ObjectP[{Model[Sample], Object[Sample]}] | _String) | (Automatic | Null), RangeP[0*Microliter, 50*Microliter, 1*Microliter] | (Automatic | Null), RangeP[30*Celsius, 90*Celsius] | (Ambient | Automatic), ObjectP[Object[Method, Gradient]] | (Automatic | New), ObjectP[Object[Method, MassAcquisition]] | (Automatic | New)}..} | Automatic
SampleTemperature
The temperature of the chamber in which the samples, Standard, and Blank are stored while waiting for the Injection.
Pattern Description: Ambient or greater than or equal to 5 degrees Celsius and less than or equal to 40 degrees Celsius.
Programmatic Pattern: RangeP[5*Celsius, 40*Celsius] | Ambient
ColumnTemperature
The temperature of the Column throughout the measurement and/or collection. If ColumnTemperature is set to Ambient, column oven temperature control is not used. Otherwise, ColumnTemperature is maintained by temperature control of the column oven.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the Gradient option or the column temperature for the sample in the InjectionTable option; otherwise, set to Ambient.
Pattern Description: Ambient or greater than or equal to 30 degrees Celsius and less than or equal to 90 degrees Celsius.
Programmatic Pattern: (RangeP[30*Celsius, 90*Celsius] | Ambient) | Automatic
Index Matches to: experiment samples
InjectionVolume
The physical quantity of sample loaded into the flow path for measurement.
Default Calculation: If InjectionTable is specified, automatically set from the Injection Volume entry for the sample. Otherwise set to 5 Microliter.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 50 microliters.
Programmatic Pattern: RangeP[0*Microliter, 50*Microliter] | Automatic
Index Matches to: experiment samples
NeedleWashSolution
The solvent used to wash the injection needle before each sample introduction.
Default Calculation: Defaults to Model[Sample, "Milli-Q water"] for IonExchange and SizeExclusion SeparationType or Model[Sample, StockSolution, "20% Methanol in MilliQ Water"] for other SeparationType.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample]
Programmatic Pattern: ObjectP[{Object[Sample], Model[Sample]}] | Automatic
Gradient
GradientA
The composition of BufferA within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientB, GradientC, and GradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic
Index Matches to: experiment samples
GradientB
The composition of BufferB within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientA, GradientC, and GradientD options such that the total amounts to 100%. If no other gradient options are specified, a Buffer B gradient of 10% to 100% over 45 minutes is used.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic
Index Matches to: experiment samples
GradientC
The composition of BufferC within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientA, GradientB, and GradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries.
Programmatic Pattern: (RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic
Index Matches to: experiment samples
GradientD
The composition of BufferD within the flow, defined for specific time points. The composition is linearly interpolated for the intervening periods between the defined time points. For example for GradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If Gradient option is specified, set from it or implicitly determined from the GradientA, GradientB, and GradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: experiment samples
FlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the gradient options. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If Gradient option is specified, automatically set from the method given in the Gradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 2 milliliters per minute or list of one or more {Time, Flow Rate} entries.
Programmatic Pattern: (RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic
Index Matches to: experiment samples
Gradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow. Specific parameters of a gradient object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the Gradient options (e.g. GradientA, GradientB, GradientC, GradientD, FlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate} entries.
Programmatic Pattern: (ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic
Index Matches to: experiment samples
Separation
MaxAcceleration
When ramping up the FlowRate of solvent through the instrument, the maximum allowed change per time in the FlowRate.
Default Calculation: For Waters instruments, automatically set to the lowest value from Max the Column, Instrument, and GuardColumn models. For other instruments, automatically set to Null.
Pattern Description: Greater than 0 milliliters per minute squared or Null.
Programmatic Pattern: (GreaterP[0*(Milliliter/Minute/Minute)] | Automatic) | Null
Mass Analysis
Calibrant
The sample with components of known mass-to-charge ratios (m/z) used to calibrate the mass spectrometer. In the chosen ion polarity mode, the calibrant should contain at least 3 masses spread over the mass range of interest.
Default Calculation: If using QTOF as the mass analyzer, set to sodium iodide for peptide samples, cesium iodide for intact protein analysis. For other types of samples, is set to cesium iodide if molecular weight is above 2000 Da, to sodium iodide if molecular weight between 1200 and 2000 Da, and to sodium formate for all others (small molecule range). If using TripleQuadrupole as the mass analyzer, this option will be set automatically based on the first IonMode: Model[Sample, "id:zGj91a71kXEO"] or Model[Sample, "id:bq9LA0JA1YJz"] for Positive and Negative, respectively.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample.
Programmatic Pattern: (ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic
SecondCalibrant
The additional sample with components of known mass-to-charge ratios (m/z) used to calibrate the mass spectrometer. In the chosen ion polarity mode, the calibrant should contain at least 3 masses spread over the mass range of interest.
Default Calculation: Set to Model[Sample, "id:zGj91a71kXEO"] or Model[Sample, "id:bq9LA0JA1YJz"] for Positive and Negative, respectively, when using TripleQuadrupole as the MassAnalyzer. Otherwise set to Null.
Pattern Description: An object of type or subtype Object[Sample] or Model[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Object[Sample], Model[Sample]}] | _String) | Automatic) | Null
Analytes
The compounds of interest that are present in the given samples, used to determine the other settings for the Mass Spectrometer (e.g. MassRange).
Default Calculation: If populated, will resolve to the user-specified Analytes field in the Object[Sample]. Otherwise, will resolve to the larger compounds in the sample, in the order of Proteins, Peptides, Oligomers, then other small molecules. Otherwise, set Null.
Pattern Description: List of one or more an object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] entries or Null.
Programmatic Pattern: ({ObjectP[IdentityModelTypes]..} | Automatic) | Null
Index Matches to: experiment samples
IonMode
Indicates if positively or negatively charged ions are analyzed.
Default Calculation: For oligomer samples of the types Peptide, DNA, and other synthetic oligomers, is automatically set to positive ion mode. For other types of samples, defaults to positive ion mode, unless the sample is acid (pH<=5 or pKa<=8).
Pattern Description: Negative or Positive.
Programmatic Pattern: IonModeP | Automatic
Index Matches to: experiment samples
MassSpectrometryMethod
The previously specified instruction(s) for the analyte ionization, selection, fragmentation, and detection.
Default Calculation: If set in the InjectionTable option, set to that; otherwise, set to New.
Pattern Description: An object of type or subtype Object[Method, MassAcquisition] or New or Null.
Programmatic Pattern: ((ObjectP[Object[Method, MassAcquisition]] | New) | Automatic) | Null
Index Matches to: experiment samples
AcquisitionWindow
The time range with respect to the chromatographic separation to conduct analyte ionization, selection/survey, optional fragmentation, and detection.
Default Calculation: Set to the entire gradient window 0 Minute to the last time point in Gradient.
Pattern Description: A span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours or list of one or more a span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours or {Automatic, Null} entries.
Programmatic Pattern: (RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour] | {(RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour] | (Automatic | Null))..}) | Automatic
Index Matches to: experiment samples
AcquisitionMode
The method by which spectra are collected. DataDependent will depend on the properties of the measured mass spectrum of the intact ions. DataIndependent will systemically scan through all of the intact ions. MS1 will focus on defined intact masses. MS1MS2 will focus on fragmented masses.
Default Calculation: Set to MS1FullScan unless DataDependent related options are set, then set to DataDependent.
Pattern Description: DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring or list of one or more DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring entries.
Programmatic Pattern: (MSAcquisitionModeP | {MSAcquisitionModeP..}) | Automatic
Index Matches to: experiment samples
Fragment
Indicates if ions should be collided with neutral gas and dissociated in order to measure the resulting product ions. Also known as tandem mass spectrometry or MS/MS (as opposed to MS).
Default Calculation: Set to True if AcquisitionMode is MS1MS2ProductIonScan, DataDependent, or DataIndependent. Set True if any of the Fragmentation related options are set (e.g. FragmentMassDetection).
Pattern Description: List of one or more True or False entries or True or False.
Programmatic Pattern: (BooleanP | {BooleanP..}) | Automatic
Index Matches to: experiment samples
MassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for intact masses. When Fragment is True, the intact ions will be selected for fragmentation.
Default Calculation: For Fragment -> False, automatically set to one of three default mass ranges according to the molecular weight of the Analytes to encompass them.
Pattern Description: DataDependent or DataIndependent or MS1FullScan, NeutralIonLoss, DataDependent, DataIndependent or PrecursorIonScan or MS1MS2ProductIonScan or SelectedIonMonitoring or SelectedIonMonitoring or MultipleReactionMonitoring or list of one or more DataDependent or DataIndependent or MS1FullScan, NeutralIonLoss, DataDependent, DataIndependent or PrecursorIonScan or MS1MS2ProductIonScan or SelectedIonMonitoring or SelectedIonMonitoring or MultipleReactionMonitoring entries.
Programmatic Pattern: ((RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | MSAnalyteGroupP) | {(RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | MSAnalyteGroupP)..}) | Automatic
Index Matches to: experiment samples
MassDetectionStepSize
Indicate the step size for mass collection in range when using TripleQuadruploe as the MassAnalyzer.
Default Calculation: This option will be set to Null if using ESI-QTOF. For ESI-QQQ, if both of the mass analyzer are in mass selection mode (SelectedIonMonitoring and MultipleReactionMonitoring mode), this option will be auto resolved to Null. In all other mass scan modes in ESI-QQQ, this option will be automatically resolved to 0.1 g/mol.
Pattern Description: Greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or list of one or more greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | {(RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
ScanTime
The duration of time allowed to pass between each spectral acquisition. When AcquisitionMode is DataDependent, this value refers to the duration for measuring spectra from the intact ions. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired.
Default Calculation: Set to 1 second unless a method is given.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries.
Programmatic Pattern: (RangeP[0.015*Second, 10*Second] | {RangeP[0.015*Second, 10*Second]..}) | Automatic
Index Matches to: experiment samples
FragmentMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for product ions. When AcquisitionMode is DataDependent|DataIndependent, all of the product ions in consideration for measurement. Null if Fragment is False.
Default Calculation: When Fragment is False, set to Null. Otherwise, 20 Gram/Mole to the maximum MassDetection.
Pattern Description: DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null or list of one or more DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null entries.
Programmatic Pattern: ((({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..}) | {({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..})..}) | Automatic) | Null
Index Matches to: experiment samples
CollisionEnergy
The voltage by which intact ions are accelerated through inert gas in order to dissociate them into measurable fragment ion species when Fragment is True. If the corresponding AcquisitionMode is DataIndependent, CollisionEnergy specifies the voltage of the low fragemention scan. Otherwise, CollisionEnergy cannot be defined simultaneously with CollisionEnergyMassProfile.
Default Calculation: Is automatically set to 40 Volt when Fragment is True, otherwise is set to Null.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 255 volts or greater than or equal to -180 volts and less than or equal to 5 volts or list of one or more A list of Single Values or Single Values or Null entries or Null.
Programmatic Pattern: (((RangeP[0.1*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {((RangeP[0.1*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {(RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt])..} | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
CollisionCellExitVoltage
Also known as the Collision Cell Exit Potential (CXP). This value focuses and accelerates the ions out of collision cell (Q2) and into 2nd mass analyzer (MS 2). This potential is tuned to ensure successful ion acceleration out of collision cell and into MS2, and can be adjusted to reach the maximal signal intensity. This option is unique to ESI-QQQ for now, and only required when Fragment ->True and/or in ScanMode that achieves tandem mass feature (PrecursorIonScan, NeutralIonLoss,MS1MS2ProductIonScan,MultipleReactionMonitoring). For non-tandem mass ScanMode (FullScan and SelectedIonMonitoring) and other massspectrometer (ESI-QTOF and MALDI-TOF), this option is resolved to Null.
Default Calculation: Is automatically set to 15 V (Positive mode) or -15 V (Negative mode) if the collision option is True and using QQQ as the mass analyzer.
Pattern Description: Greater than or equal to -55 volts and less than or equal to 55 volts or list of one or more greater than or equal to -55 volts and less than or equal to 55 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[-55*Volt, 55*Volt] | {(RangeP[-55*Volt, 55*Volt] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
DwellTime
The duration of time for which spectra are acquired at the specific mass detection value for SelectedIonMonitoring and MultipleReactionMonitoring mode in ESI-QQQ.
Default Calculation: Is automatically set to 200 microsecond if StandardAcquisition is in SelectedIonMonitoring or MultipleReactionMonitoring mode. Otherwise is set to Null.
Pattern Description: Greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more A list of Single Values or Single Values or Null entries or Null.
Programmatic Pattern: ((RangeP[5*Millisecond, 2000*Millisecond] | {(RangeP[5*Millisecond, 2000*Millisecond] | {RangeP[5*Millisecond, 2000*Millisecond]..} | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
NeutralLoss
A neutral loss scan is performed on ESI-QQQ instrument by scanning the sample through the first quadrupole (Q1). The ions are then fragmented in the collision cell. The second mass analyzer is then scanned with a fixed offset to MS1. This option represents the value of this offset.
Default Calculation: Is set to 500 g/mol if using NeutralIonLoss as AcquisitionMode, and is Null in other modes.
Pattern Description: Greater than 0 grams per mole or list of one or more greater than 0 grams per mole or Null entries or Null.
Programmatic Pattern: ((GreaterP[0*(Gram/Mole)] | {(GreaterP[0*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
MultipleReactionMonitoringAssays
In ESI-QQQ, the ion corresponding to the compound of interest is targeted with subsequent fragmentation of that target ion to produce a range of daughter ions. One (or more) of these fragment daughter ions can be selected for quantitation purposes. Only compounds that meet both these criteria, i.e. specific parent ion and specific daughter ions corresponding to the mass of the molecule of interest are detected within the mass spectrometer. The mass assays (MS1/MS2 mass value combinations) for each scan, along with the CollisionEnergy and DwellTime (length of time of each scan).
Default Calculation: Is set based on MassDetection, CollisionEnergy, DwellTime and FragmentMassDetection.
Pattern Description: List of one or more Individual Multiple Reaction Monitoring Assay or {Multiple Reaction Monitoring Assays} or None entries or Null.
Programmatic Pattern: (({({{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..} | {{{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..}} | (Null | {Null}))..} | Null) | Automatic) | Null
Index Matches to: experiment samples
CollisionEnergyMassProfile
The relationship of collision energy with the MassDetection. If the corresponding AcquisitionMode is DataIndependent, this span specifies the minimum and maximum values for the linear voltage ramp of the high fragmentation scan. If the corresponding AcquisitionMode is DataDependent, this span specifies the minimum and maximum values for the low energy voltage ramp. Otherwise, it should be Null.
Default Calculation: Set to CollisionEnergyMassScan if defined; otherwise, set to Null.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or list of one or more a span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | {(RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
CollisionEnergyMassScan
The collision energy profile at the end of the scan from CollisionEnergy or CollisionEnergyScanProfile, as related to analyte mass.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or list of one or more a span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | {(RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
SurveyChargeStateExclusion
Indicates if redundant ions that differ by ionic charge (+1/-1, +2/-2, etc.) should be excluded and if ChargeState exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the ChargeState options are set; otherwise, False.
Pattern Description: List of one or more True or False or Null entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {(BooleanP | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
SurveyIsotopeExclusion
Indicates if redundant ions that differ by isotopic mass (e.g. 1, 2 Gram/Mole) should be excluded and if MassIsotope exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the IsotopeExclusion options are set; otherwise, False.
Pattern Description: List of one or more True or False or Null entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {(BooleanP | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
Ionization
ESICapillaryVoltage
The absolute voltage applied to the tip of the stainless steel capillary tubing in order to produce charged droplets. Adjust this voltage to maximize sensitivity. Most compounds are optimized between 0.5 and 3.2 kV in ESI positive ion mode and 0.5 and 2.6 in ESI negative ion mode, but can be altered according to sample type. For low flow applications, best sensitivity will be achieved with a relatively high value in ESI positive (e.g. 3.0 kV), for standard flow UPLC a value of 0.5 kV is typically best for maximum sensitivity.
Default Calculation: Is automatically set according to the flow rate (0-0.02 ml/min -> 3.0 kV, 0.02-0.1 ml/min -> 1.5 kV, >0.1 ml/min -> 0.5 kV).
Pattern Description: Greater than or equal to -4 kilovolts and less than or equal to 5 kilovolts.
Programmatic Pattern: RangeP[-4*Kilovolt, 5*Kilovolt] | Automatic
Index Matches to: experiment samples
DeclusteringVoltage
The voltage offset between the ion block (the reduced pressure chamber of the source block) and the stepwave ion guide (the optics before the quadrupole mass analyzer). This voltage attracts charged ions in the spray being produced from the capillary tip into the ion block leading into the mass spectrometer. This voltage is typically set to 25-100 V and its tuning has little effect on sensitivity compared to other options (e.g. StepwaveVoltage).
Default Calculation: Is automatically set to any specified MassAcquisition method; otherwise, set to 40 Volt.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts.
Programmatic Pattern: RangeP[0.1*Volt, 150*Volt] | Automatic
Index Matches to: experiment samples
StepwaveVoltage
The voltage offset between the 1st and 2nd stage of the ion guide which leads ions coming from the sample cone towards the quadrupole mass analyzer. This voltage normally optimizes between 25 and 150 V and should be adjusted for sensitivity depending on compound and charge state. For multiply charged species it is typically set to to 40-50 V, and higher for singly charged species. In general higher cone voltages (120-150 V) are needed for larger mass ions such as intact proteins and monoclonal antibodies. It also has greatest effect on in-source fragmentation and should be decreased if in-source fragmentation is observed but not desired. This is an unique option for QTOF as the massanalyzer.
Default Calculation: Is automatically set according to the sample type (proteins, antibodies and analytes with MW > 2000 -> 120 V, DNA and synthetic nucleic acid oligomers -> 100 V, all others (including peptides and small molecules) -> 40 V).
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 200 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 200*Volt] | Automatic) | Null
Index Matches to: experiment samples
SourceTemperature
The temperature setting of the source block. Heating the source block discourages condensation and decreases solvent clustering in the reduced vacuum region of the source. This temperature setting is flow rate and sample dependent. Typical values are between 60 to 120 Celsius. For thermally labile analytes, a lower temperature setting is recommended.
Default Calculation: Is automatically set according to the flow rate (0-0.02 ml/min -> 100 Celsius, 0.02-0.3 ml/min -> 120 Celsius, >0.301 -> 150 Celsius).
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius.
Programmatic Pattern: RangeP[25*Celsius, 150*Celsius] | Automatic
Index Matches to: experiment samples
DesolvationTemperature
The temperature setting for the ESI desolvation heater that controls the nitrogen gas temperature used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to DesolvationGasFlow, this setting is dependent on solvent flow rate and composition. A typical range is 150 to 650 Celsius.
Default Calculation: Is automatically set according to the flow rate (0-0.02 ml/min -> 200 Celsius, 0.02-0.1 ml/min -> 350 Celsius, 0.101-0.3 -> 450 Celsius, 0.301->0.5 ml/min -> 500 Celsius, >0.500 ml/min -> 600 Celsius).
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 650 degrees Celsius.
Programmatic Pattern: RangeP[20*Celsius, 650*Celsius] | Automatic
Index Matches to: experiment samples
DesolvationGasFlow
The rate at which nitrogen gas is flowed around the ESI capillary. It is used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to DesolvationTemperature, this setting is dependent on solvent flow rate and composition. Higher desolvation gas flows usually result in increased sensitivity, but too high values can cause spray instability. Typical values are between 300 and 1200 L/h.
Default Calculation: Is automatically set according to the flow rate (0-0.02 ml/min -> 600 L/h, 0.02-0.3 ml/min -> 800 L/h, 0.301-0.500 ml/min -> 1000 L/h, >0.500 ml/min -> 1200 L/h).
Pattern Description: Greater than or equal to 55 liters per hour and less than or equal to 1200 liters per hour or greater than or equal to 0 pounds
‐force per inch squared and less than or equal to 85 pounds
‐force per inch squared.
Programmatic Pattern: (RangeP[55*(Liter/Hour), 1200*(Liter/Hour)] | RangeP[0*PSI, 85*PSI]) | Automatic
Index Matches to: experiment samples
ConeGasFlow
The nitrogen gas flow ejected around the sample inlet cone (the spherical metal plate acting as a first gate between the sprayer and the reduced pressure chamber, the ion block). This gas flow is used to minimize the formation of solvent ion clusters. It also helps reduce adduct ions and directing the spray into the ion block while keeping the sample cone clean. The same parameter is referred to as Curtain Gas Pressure for ESI-QQQ. Typical values are between 0 and 150 L/h for ESI-QTOF or 20 to 55 PSI for ESI-QQQ.
Default Calculation: Is automatically set to 50 Liter/Hour for ESI-QTOF and 50 PSI for ESI-QQQ, and is set to Null in MALDI-TOF. Is not recommended to set to a smaller value of 40 PSI in ESI-QQQ, due to potential deposition of the sample inside the instrument that will lead to contamination.
Pattern Description: Greater than or equal to 0 liters per hour and less than or equal to 300 liters per hour or greater than or equal to 20 pounds
‐force per inch squared and less than or equal to 55 pounds
‐force per inch squared.
Programmatic Pattern: (RangeP[0*(Liter/Hour), 300*(Liter/Hour)] | RangeP[20*PSI, 55*PSI]) | Automatic
Index Matches to: experiment samples
IonGuideVoltage
This option (also known as Entrance Potential (EP)) is a unique option of ESI-QQQ. This parameter indicates electric potential applied to the Ion Guide in ESI-QQQ, which guides and focuses the ions through the high-pressure ion guide region.
Default Calculation: Is automatically set to 10 V for positive ions, or
–10 V for negative ions in ESI-QQQ, and can be changed between 2-15 V in both positive and negative mode. This value is set to Null in ESI-QTOF.
Pattern Description: Greater than or equal to -15 volts and less than or equal to -2 volts or greater than or equal to 2 volts and less than or equal to 15 volts or Null.
Programmatic Pattern: ((RangeP[-15*Volt, -2*Volt] | RangeP[2*Volt, 15*Volt]) | Automatic) | Null
Index Matches to: experiment samples
StandardIonGuideVoltage
This option (also known as Entrance Potential (EP)) is a unique option of ESI-QQQ. This parameter indicates electric potential applied to the Ion Guide in ESI-QQQ, which guides and focuses the ions through the high-pressure ion guide region.
Default Calculation: Is automatically set to the first IonGuideVoltage.
Pattern Description: Greater than or equal to -15 volts and less than or equal to -2 volts or greater than or equal to 2 volts and less than or equal to 15 volts or Null.
Programmatic Pattern: ((RangeP[-15*Volt, -2*Volt] | RangeP[2*Volt, 15*Volt]) | Automatic) | Null
Index Matches to: Standard
BlankIonGuideVoltage
This option (also known as Entrance Potential (EP)) is a unique option of ESI-QQQ. This parameter indicates electric potential applied to the Ion Guide in ESI-QQQ, which guides and focuses the ions through the high-pressure ion guide region.
Default Calculation: Is automatically set to the first IonGuideVoltage.
Pattern Description: Greater than or equal to -15 volts and less than or equal to -2 volts or greater than or equal to 2 volts and less than or equal to 15 volts or Null.
Programmatic Pattern: ((RangeP[-15*Volt, -2*Volt] | RangeP[2*Volt, 15*Volt]) | Automatic) | Null
ColumnPrimeIonGuideVoltage
This option (also known as Entrance Potential (EP)) is a unique option of ESI-QQQ. This parameter indicates electric potential applied to the Ion Guide in ESI-QQQ, which guides and focuses the ions through the high-pressure ion guide region.
Default Calculation: Is automatically set to first IonGuideVoltage.
Pattern Description: Greater than or equal to -15 volts and less than or equal to -2 volts or greater than or equal to 2 volts and less than or equal to 15 volts or Null.
Programmatic Pattern: ((RangeP[-15*Volt, -2*Volt] | RangeP[2*Volt, 15*Volt]) | Automatic) | Null
ColumnFlushIonGuideVoltage
This option (also known as Entrance Potential (EP)) is a unique option of ESI-QQQ. This parameter indicates electric potential applied to the Ion Guide in ESI-QQQ, which guides and focuses the ions through the high-pressure ion guide region.
Default Calculation: Is automatically set to first IonGuideVoltage.
Pattern Description: Greater than or equal to -15 volts and less than or equal to -2 volts or greater than or equal to 2 volts and less than or equal to 15 volts or Null.
Programmatic Pattern: ((RangeP[-15*Volt, -2*Volt] | RangeP[2*Volt, 15*Volt]) | Automatic) | Null
Data Dependent Acquisition
FragmentScanTime
The duration of the spectral scanning for each fragmentation of an intact ion when AcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to the same value as ScanTime if AcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {(RangeP[0.015*Second, 10*Second] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
AcquisitionSurvey
The number of intact ions to consider for fragmentation and product ion measurement in every measurement cycle when AcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to 10 if AcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 30 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 30 in increments of 1 or Null entries or Null.
Programmatic Pattern: ((RangeP[1, 30, 1] | {(RangeP[1, 30, 1] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
MinimumThreshold
The minimum number of total ions detected within ScanTime durations needed to trigger the start of data dependent acquisition when AcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to (100000/Second)*ScanTime if AcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or list of one or more greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null entries or Null.
Programmatic Pattern: ((RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | {(RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
AcquisitionLimit
The maximum number of total ions for a specific intact ion when AcquisitionMode is set to DataDependent. When this value is exceeded, acquisition will switch to fragmentation of the next candidate ion.
Default Calculation: Automatically inherited from supplied method if AcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or list of one or more greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null entries or Null.
Programmatic Pattern: ((RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | {(RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
CycleTimeLimit
The maximum possible computed duration of all of the scans for the intact and fragmentation measurements when AcquisitionMode is set to DataDependent.
Default Calculation: Calculated from the AcquisitionSurvey, ScanTime, and FragmentScanTime.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 20000*Second] | {(RangeP[0.015*Second, 20000*Second] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
ExclusionDomain
The time range when the ExclusionMasses are omitted in the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire AcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or Null entries or list of one or more list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full | Null)..} | {{(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}..}) | Automatic) | Null
Index Matches to: experiment samples
ExclusionMass
The intact ions (Target Mass) to omit. When set to All, the mass is excluded for the entire ExclusionDomain. When set to Once, the mass is excluded in the first survey appearance, but considered for consequent ones.
Default Calculation: If any ExclusionMode-related options are set (e.g. ExclusionMassTolerance), a target mass of the first Analyte (if not in InclusionMasses) is chosen and retention time is set to 0*Minute.
Pattern Description: List of one or more list of one or more {Mode, Target Mass} entries entries or list of one or more {Mode, Target Mass} or Null entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | {({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | Null)..} | {{{All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]}..}..}) | Automatic) | Null
Index Matches to: experiment samples
ExclusionMassTolerance
The range above and below each ion in ExclusionMasses to consider for omission when ExclusionMass is set to All or Once.
Default Calculation: If ExclusionMass -> All or Once, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
ExclusionRetentionTimeTolerance
The range of time above and below the ExclusionDomain to consider for exclusion.
Default Calculation: If ExclusionMass and ExclusionDomain options are set, this is set to 10 seconds; otherwise, Null.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 3600 seconds or list of one or more greater than or equal to 0 seconds and less than or equal to 3600 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0*Second, 3600*Second] | {(RangeP[0*Second, 3600*Second] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
InclusionDomain
The time range when InclusionMass applies in the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire AcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or Null entries or list of one or more list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full | Null)..} | {{(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}..}) | Automatic) | Null
Index Matches to: experiment samples
InclusionMass
The ions (Target Mass) to prioritize during the survey scan for further fragmentation when AcquisitionMode is DataDependent. When the Mode is Only, the InclusionMass will solely be considered for surveys. When Mode is Preferential, the InclusionMass will be prioritized for survey.
Default Calculation: When InclusionMode Only or Preferential, an entry mass is added based on the mass of the most salient analyte of the sample.
Pattern Description: List of one or more list of one or more {Mode, Target Mass} entries entries or list of one or more {Mode, Target Mass} or Null entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | {({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | Null)..} | {{{Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]}..}..}) | Automatic) | Null
Index Matches to: experiment samples
InclusionCollisionEnergy
The overriding collision energy value that can be applied to the InclusionMass. Null will default to the CollisionEnergy and related options.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 volts and less than or equal to 255 volts or list of one or more greater than or equal to 0 volts and less than or equal to 255 volts or Null entries or list of one or more list of one or more greater than or equal to 0 volts and less than or equal to 255 volts entries entries or Null.
Programmatic Pattern: ((RangeP[0*Volt, 255*Volt] | {(RangeP[0*Volt, 255*Volt] | Null)..} | {{RangeP[0*Volt, 255*Volt]..}..}) | Automatic) | Null
Index Matches to: experiment samples
InclusionDeclusteringVoltage
The overriding source voltage value that can be applied to the InclusionMass. Null will default to the DeclusteringVoltage option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts or Null entries or list of one or more list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts entries entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 150*Volt] | {(RangeP[0.1*Volt, 150*Volt] | Null)..} | {{RangeP[0.1*Volt, 150*Volt]..}..}) | Automatic) | Null
Index Matches to: experiment samples
InclusionChargeState
The maximum charge state of the InclusionMass to also consider for inclusion. For example, if this is set to 3 and the polarity is Positive, then +1,+2,+3 charge states will be considered as well.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 or Null entries or list of one or more list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 entries entries or Null.
Programmatic Pattern: ((RangeP[0, 6, 1] | {(RangeP[0, 6, 1] | Null)..} | {{RangeP[0, 6, 1]..}..}) | Automatic) | Null
Index Matches to: experiment samples
InclusionScanTime
The overriding scan time duration that can be applied to the InclusionMass for the consequent fragmentation. Null will default to the FragmentScanTime option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null entries or list of one or more list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {(RangeP[0.015*Second, 10*Second] | Null)..} | {{RangeP[0.015*Second, 10*Second]..}..}) | Automatic) | Null
Index Matches to: experiment samples
InclusionMassTolerance
The range above and below each ion in InclusionMasses to consider for prioritization. For example, if set to 0.5 Gram/Mole, the total range is 1 Gram/Mole.
Default Calculation: Set to 0.5 Gram/Mole if InclusionMass is given; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
ChargeStateExclusionLimit
The number of ions to survey first with exclusion by ionic state. For example, if AcquisitionSurvey is 10 and this option is 5, then 5 ions will be surveyed with charge-state exclusion. For candidate ions of rank 6 to 10, no exclusion will be performed.
Default Calculation: Inherited from any supplied method; otherwise, set the same to AcquisitionSurvey, if any ChargeState option is set.
Pattern Description: Greater than or equal to 0 and less than or equal to 30 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 30 in increments of 1 or Null entries or Null.
Programmatic Pattern: ((RangeP[0, 30, 1] | {(RangeP[0, 30, 1] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
ChargeStateExclusion
The specific ionic states of intact ions to redundantly exclude from the survey for further fragmentation/acquisition. 1 refers to +1/-1, 2 refers to +2/-2, etc.
Default Calculation: When SurveyChargeStateExclusion is True, set to {1,2}; otherwise, Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 entries or Null entries or Null.
Programmatic Pattern: ((RangeP[1, 6, 1] | {(RangeP[1, 6, 1] | {RangeP[1, 6, 1]..} | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
ChargeStateMassTolerance
The range of m/z to consider for exclusion by ionic state property when SurveyChargeStateExclusion is True.
Default Calculation: When SurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
IsotopicExclusion
The m/z difference between monoisotopic ions as a criterion for survey exclusion.
Default Calculation: When SurveyIsotopeExclusion is True, set to 1 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole entries or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]..} | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
IsotopeRatioThreshold
The minimum relative magnitude between monoisotopic ions in order to be considered an isotope for exclusion.
Default Calculation: When SurveyIsotopeExclusion is True, set to 0.1; otherwise, Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 entries or Null entries or Null.
Programmatic Pattern: ((RangeP[0, 1] | {(RangeP[0, 1] | {RangeP[0, 1]..} | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
IsotopeDetectionMinimum
The acquisition rate of a given intact mass to consider for isotope exclusion in the survey.
Default Calculation: When SurveyIsotopeExclusion is True, set to 10 1/Second; otherwise, Null.
Pattern Description: Greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds entries or Null entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(1/Second)] | {(GreaterEqualP[0*(1/Second)] | {GreaterEqualP[0*(1/Second)]..} | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
IsotopeMassTolerance
The range of m/z around a mass to consider for exclusion. This applies for both ChargeState and mass shifted Isotope. If set to 0.5 Gram/Mole, then the total range should be 1 Gram/Mole.
Default Calculation: When SurveyIsotopeExclusion or SurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
IsotopeRatioTolerance
The range of relative magnitude around IsotopeRatio to consider for isotope exclusion.
Default Calculation: If SurveyIsotopeExclusion is True, set to 30*Percent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more greater than or equal to 0 percent and less than or equal to 100 percent or Null entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {(RangeP[0*Percent, 100*Percent] | Null)..}) | Automatic) | Null
Index Matches to: experiment samples
Detector Parameters
AbsorbanceWavelength
The wavelength of light passed through the flow cell for the PhotoDiodeArray (PDA) Detector. A 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: When conducting absorbance measurement, set to All.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 500*Nanometer] | All | RangeP[190*Nanometer, 500*Nanometer] ;; RangeP[200*Nanometer, 500*Nanometer]) | Automatic) | Null
Index Matches to: experiment samples
WavelengthResolution
The increment of wavelength for the range of light passed through the flow for absorbance measurement with the PhotoDiodeArray detector.
Default Calculation: Automatically set to 2.4 Nanometer. Set to Null if AbsorbanceWavelength is a singleton value.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[1.2*Nanometer, 12.*Nanometer] | Automatic) | Null
Index Matches to: experiment samples
UVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the sample for PhotoDiodeArray detectors.
Default Calculation: Automatically set to False for Instruments with PDA detectors; otherwise, resolves to Null.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: experiment samples
AbsorbanceSamplingRate
The number of times an absorbance measurement is made per second by the detector on the selected instrument. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: Automatically set to 20/Second for Instruments with PhotoDiodeArray (PDA) detectors; otherwise, resolves to Null.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second), 1*(1/Second)] | Automatic) | Null
Index Matches to: experiment samples
Standard
Standard
The reference compound(s) injected into the instrument, often used for quantification or to check internal measurement consistency.
Default Calculation: If any other Standard option is specified, automatically set based on the SeparationMode option. If InjectionTable is specified, set from the specified Standard entries in the InjectionTable.
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
Index Matches to: Standard
StandardInjectionVolume
The physical quantity of Standard sample loaded into the flow path on the selected instrument along with the mobile phase onto the stationary phase.
Default Calculation: Automatically set equal to the first entry in InjectionVolume.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 500 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 500*Microliter] | Automatic) | Null
Index Matches to: Standard
StandardFrequency
The frequency at which Standard measurements will be inserted between the experiment samples.
Default Calculation: If injectionTable is given, automatically set to Null and the sequence of Standards specified in the InjectionTable will be used in the experiment. If any other Standard option is specified, automatically set to FirstAndLast.
Pattern Description: Greater than 0 in increments of 1 or None, First, Last, FirstAndLast, or GradientChange or Null.
Programmatic Pattern: (((None | First | Last | FirstAndLast | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
StandardColumnTemperature
The temperature of the column when the Standard gradient and measurement are run. If StandardColumnTemperature is set to Ambient, column oven temperature control is not used. Otherwise, StandardColumnTemperature is maintained by temperature control of the column oven.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the StandardGradient option or the column temperature for the sample in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 30 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[30*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
Index Matches to: Standard
StandardGradientA
The composition of BufferA within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientB, StandardGradientC, and StandardGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradientB
The composition of BufferB within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientA, StandardGradientC, and StandardGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradientC
The composition of BufferC within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientA, StandardGradientB, and StandardGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradientD
The composition of BufferD within the flow, defined for specific time points for Standard measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for StandardGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If StandardGradient option is specified, set from it or implicitly determined from the StandardGradientA, StandardGradientB, and StandardGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
Index Matches to: Standard
StandardFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the StandardGradient options during Standard measurement. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If StandardGradient option is specified, automatically set from the method given in the StandardGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 2 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
Index Matches to: Standard
StandardGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow for Standard measurement. Specific parameters of a gradient object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the StandardGradient options (e.g. StandardGradientA, StandardGradientB, StandardGradientC, StandardGradientD, StandardFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
Index Matches to: Standard
StandardAnalytes
The compounds of interest that are present in the given Standard, used to determine the other settings for the Mass Spectrometer (e.g. MassRange).
Default Calculation: If populated, will resolve to the user-specified Analytes field in the Object[Sample]. Otherwise, will resolve to the larger compounds in the sample, in the order of Proteins, Peptides, Oligomers, then other small molecules; otherwise, set Null.
Pattern Description: List of one or more an object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] entries or Null.
Programmatic Pattern: ({ObjectP[IdentityModelTypes]..} | Automatic) | Null
Index Matches to: Standard
StandardIonMode
Indicates if positively or negatively charged ions are analyzed for the Standard.
Default Calculation: Set to the first IonMode for an analyte input sample.
Pattern Description: Negative or Positive or Null.
Programmatic Pattern: (IonModeP | Automatic) | Null
Index Matches to: Standard
StandardMassSpectrometryMethod
The previously specified instruction(s) for the Standard ionization, selection, fragmentation, and detection.
Default Calculation: If Standard samples exist and MassSpectrometryMethod is specified, then set to the first available StandardMassSpectrometryMethod.
Pattern Description: An object of type or subtype Object[Method, MassAcquisition] or New or Null.
Programmatic Pattern: ((ObjectP[Object[Method, MassAcquisition]] | New) | Automatic) | Null
Index Matches to: Standard
StandardESICapillaryVoltage
The absolute voltage applied to the tip of the stainless steel capillary tubing in order to produce charged droplets. Adjust this voltage to maximize sensitivity. Most compounds are optimized between 0.5 and 3.2 kV in ESI positive ion mode and 0.5 and 2.6 in ESI negative ion mode, but can be altered according to sample type. For low flow applications, best sensitivity will be achieved with a relatively high value in ESI positive (e.g. 3.0 kV), for standard flow UPLC a value of 0.5 kV is typically best for maximum sensitivity.
Default Calculation: Is automatically set to the first ESICapillaryVoltage.
Pattern Description: Greater than or equal to -4 kilovolts and less than or equal to 5 kilovolts or Null.
Programmatic Pattern: (RangeP[-4*Kilovolt, 5*Kilovolt] | Automatic) | Null
Index Matches to: Standard
StandardDeclusteringVoltage
The voltage offset between the ion block (the reduced pressure chamber of the source block) and the stepwave ion guide (the optics before the quadrupole mass analyzer). This voltage attracts charged ions in the spray being produced from the capillary tip into the ion block leading into the mass spectrometer. This voltage is typically set to 25-100 V and its tuning has little effect on sensitivity compared to other options (e.g. StandardStepwaveVoltage).
Default Calculation: Is automatically set to any specified MassAcquisition method; otherwise, set to 40 Volt.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 150*Volt] | Automatic) | Null
Index Matches to: Standard
StandardStepwaveVoltage
The voltage offset between the 1st and 2nd stage of the stepwave ion guide which leads ions coming from the sample cone towards the quadrupole mass analyzer. This voltage normally optimizes between 25 and 150 V and should be adjusted for sensitivity depending on compound and charge state. For multiply charged species it is typically set to to 40-50 V, and higher for singly charged species. In general higher cone voltages (120-150 V) are needed for larger mass ions such as intact proteins and monoclonal antibodies. It also has greatest effect on in-source fragmentation and should be decreased if in-source fragmentation is observed but not desired.
Default Calculation: Is automatically set to the first StepwaveVoltage.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 200 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 200*Volt] | Automatic) | Null
Index Matches to: Standard
StandardSourceTemperature
The temperature setting of the source block. Heating the source block discourages condensation and decreases solvent clustering in the reduced vacuum region of the source. This temperature setting is flow rate and sample dependent. Typical values are between 60 to 120 Celsius. For thermally labile analytes, a lower temperature setting is recommended.
Default Calculation: Is automatically set to the first SourceTemperature.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 150*Celsius] | Automatic) | Null
Index Matches to: Standard
StandardDesolvationTemperature
The temperature setting for the ESI desolvation heater that controls the nitrogen gas temperature used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to StandardDesolvationGasFlow, this setting is dependent on solvent flow rate and composition. A typical range is from 150 to 650 Celsius.
Default Calculation: Is automatically set to the first DesolvationTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 650 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 650*Celsius] | Automatic) | Null
Index Matches to: Standard
StandardDesolvationGasFlow
The rate at which nitrogen gas is flowed around the ESI capillary. It is used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to StandardDesolvationTemperature, this setting is dependent on solvent flow rate and composition. Higher desolvation temperatures usually result in increased sensitivity, but too high values can cause spray instability. Typical values are between 300 to 1200 L/h.
Default Calculation: Is automatically set to the first DesolvationGasFlow.
Pattern Description: Greater than or equal to 55 liters per hour and less than or equal to 1200 liters per hour or greater than or equal to 0 pounds
‐force per inch squared and less than or equal to 85 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[55*(Liter/Hour), 1200*(Liter/Hour)] | RangeP[0*PSI, 85*PSI]) | Automatic) | Null
Index Matches to: Standard
StandardConeGasFlow
The rate at which nitrogen gas is flowed around the sample inlet cone (the spherical metal plate acting as a first gate between the sprayer and the reduced pressure chamber, the ion block). This gas flow is used to minimize the formation of solvent ion clusters. It also helps reduce adduct ions and directing the spray into the ion block while keeping the sample cone clean. Typical values are between 0 and 150 L/h.
Default Calculation: Is automatically set to the first ConeGasFlow.
Pattern Description: Greater than or equal to 0 liters per hour and less than or equal to 300 liters per hour or greater than or equal to 30 pounds
‐force per inch squared and less than or equal to 55 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[0*(Liter/Hour), 300*(Liter/Hour)] | RangeP[30*PSI, 55*PSI]) | Automatic) | Null
Index Matches to: Standard
StandardAcquisitionWindow
The time range with respect to the chromatographic separation to conduct standard analyte ionization, selection/survey, optional fragmentation, and detection.
Default Calculation: Set to the entire gradient window 0 Minute to the last time point in StandardGradient.
Pattern Description: A span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours or list of one or more a span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours entries or Null.
Programmatic Pattern: ((RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour] | {(Alternatives[RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour]])..}) | Automatic) | Null
Index Matches to: Standard
StandardAcquisitionMode
The method by which spectra are collected. DataDependent will depend on the properties of the measured mass spectrum of the intact ions. DataIndependent will systemically scan through all of the intact ions. MS1 will focus on defined intact masses. MS1MS2ProductIonScan will focus on fragmented masses.
Default Calculation: Set to MS1FullScan unless DataDependent related options are set, then set to DataDependent.
Pattern Description: DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring or list of one or more DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring entries or Null.
Programmatic Pattern: ((MSAcquisitionModeP | {MSAcquisitionModeP..}) | Automatic) | Null
Index Matches to: Standard
StandardFragment
Indicates if ions should be collided with neutral gas and dissociated in order to measure the resulting product ions. Also known as tandem mass spectrometry or MS/MS (as opposed to MS).
Default Calculation: Set to True if StandardAcquisitionMode is MS1MS2ProductIonScan, DataDependent, or DataIndependent. Set True if any of the Fragmentation related options are set (e.g. StandardFragmentMassDetection).
Pattern Description: List of one or more True or False entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {BooleanP..}) | Automatic) | Null
Index Matches to: Standard
StandardMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for intact masses. When StandardFragment is True, the intact ions will be selected for fragmentation.
Default Calculation: For StandardFragment -> False, automatically set to one of three default mass ranges according to the molecular weight of the StandardAnalytes to encompass them.
Pattern Description: All or Range or Single or Specific List or list of one or more All or Range or Single or Specific List entries or Null.
Programmatic Pattern: (((RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | MSAnalyteGroupP) | {(RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | MSAnalyteGroupP)..}) | Automatic) | Null
Index Matches to: Standard
StandardScanTime
The duration of time allowed to pass between each spectral acquisition. When StandardAcquisitionMode is DataDependent, this value refers to the duration for measuring spectra from the intact ions. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired.
Default Calculation: Set to 0.2 seconds unless a method is given.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {RangeP[0.015*Second, 10*Second]..}) | Automatic) | Null
Index Matches to: Standard
StandardFragmentMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for product ions. When StandardAcquisitionMode is DataDependent|DataIndependent, all of the product ions in consideration for measurement. Null if StandardFragment is False.
Default Calculation: When StandardFragment is False, set to Null. Otherwise, 20 Gram/Mole to the maximum StandardMassDetection.
Pattern Description: DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null or list of one or more DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null entries.
Programmatic Pattern: ((({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..}) | {({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..})..}) | Automatic) | Null
Index Matches to: Standard
StandardCollisionEnergy
The voltage by which intact ions are accelerated through inert gas in order to dissociate them into measurable fragment ion species when StandardFragment is True. StandardCollisionEnergy cannot be defined simultaneously with StandardCollisionEnergyMassProfile.
Default Calculation: Is automatically set to 40 Volt when StandardFragment is True, otherwise is set to Null.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 255 volts or greater than or equal to -180 volts and less than or equal to 5 volts or list of one or more A list of Single Values or Single Values or Null entries or Null.
Programmatic Pattern: (((RangeP[0.1*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {((RangeP[0.1*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {(RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt])..} | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardCollisionEnergyMassProfile
The relationship of collision energy with the StandardMassDetection.
Default Calculation: Set to StandardCollisionEnergyMassScan if defined; otherwise, set to Null.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or list of one or more a span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | {(RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardCollisionEnergyMassScan
The collision energy profile at the end of the scan from StandardCollisionEnergy or StandardCollisionEnergyScanProfile, as related to analyte mass.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or list of one or more a span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | {(RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardFragmentScanTime
The duration of the spectral scanning for each fragmentation of an intact ion when StandardAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to the same value as ScanTime if StandardAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {(RangeP[0.015*Second, 10*Second] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardAcquisitionSurvey
The number of intact ions to consider for fragmentation and product ion measurement in every measurement cycle when StandardAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to 10 if StandardAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 30 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 30 in increments of 1 or Null entries or Null.
Programmatic Pattern: ((RangeP[1, 30, 1] | {(RangeP[1, 30, 1] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardMinimumThreshold
The minimum number of total ions detected within ScanTime durations needed to trigger the start of data dependent acquisition when StandardAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to (100000/Second)*ScanTime if StandardAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or list of one or more greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null entries or Null.
Programmatic Pattern: ((RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | {(RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardAcquisitionLimit
The maximum number of total ions for a specific intact ion when StandardAcquisitionMode is set to DataDependent. When this value is exceeded, acquisition will switch to fragmentation of the next candidate ion.
Default Calculation: Automatically inherited from supplied method if StandardAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or list of one or more greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null entries or Null.
Programmatic Pattern: ((RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | {(RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardCycleTimeLimit
The maximum possible computed duration of all of the scans for the intact and fragmentation measurements when StandardAcquisitionMode is set to DataDependent.
Default Calculation: Calculated from the StandardAcquisitionSurvey, StandardScanTime, and StandardFragmentScanTime.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 20000*Second] | {(RangeP[0.015*Second, 20000*Second] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardExclusionDomain
The time range when the StandardExclusionMasses are omitted in the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire StandardAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or Null entries or list of one or more list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full | Null)..} | {{(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}..}) | Automatic) | Null
Index Matches to: Standard
StandardExclusionMass
The intact ions (Target Mass) to omit. When set to All, the mass is excluded for the entire StandardExclusionDomain. When set to Once, the mass is excluded in the first survey appearance, but considered for consequent ones.
Default Calculation: If any StandardExclusionMode-related options are set (e.g. StandardExclusionMassTolerance), a target mass of the first Analyte (if not in StandardInclusionMasses) is chosen and retention time is set to 0*Minute.
Pattern Description: List of one or more list of one or more {Mode, Target Mass} entries entries or list of one or more {Mode, Target Mass} or Null entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | {({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | Null)..} | {{{All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]}..}..}) | Automatic) | Null
Index Matches to: Standard
StandardExclusionMassTolerance
The range above and below each ion in StandardExclusionMasses to consider for omission when StandardExclusionMass is All or Once.
Default Calculation: If StandardExclusionMass -> All or Once, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardExclusionRetentionTimeTolerance
The range of time above and below the StandardExclusionDomain to consider for exclusion.
Default Calculation: If StandardExclusionMass and StandardExclusionDomain options are set, this is set to 10 seconds; otherwise, Null.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 3600 seconds or list of one or more greater than or equal to 0 seconds and less than or equal to 3600 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0*Second, 3600*Second] | {(RangeP[0*Second, 3600*Second] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardInclusionDomain
The time range when StandardInclusionMass applies with respect to the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire StandardAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or Null entries or list of one or more list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full | Null)..} | {{(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}..}) | Automatic) | Null
Index Matches to: Standard
StandardInclusionMass
The ions (Target Mass) to prioritize during the survey scan for further fragmentation when StandardAcquisitionMode is DataDependent. StandardInclusionMass set to Only will solely be considered for surveys. When Mode is Preferential, the InclusionMass will be prioritized for survey.
Default Calculation: When StandardInclusionMode Only or Preferential, an entry mass is added based on the mass of the most salient analyte of the sample.
Pattern Description: List of one or more list of one or more {Mode, Target Mass} entries entries or list of one or more {Mode, Target Mass} or Null entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | {({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | Null)..} | {{{Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]}..}..}) | Automatic) | Null
Index Matches to: Standard
StandardInclusionCollisionEnergy
The overriding collision energy value that can be applied to the StandardInclusionMass. Null will default to the StandardCollisionEnergy option and related options.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 volts and less than or equal to 255 volts or list of one or more greater than or equal to 0 volts and less than or equal to 255 volts or Null entries or list of one or more list of one or more greater than or equal to 0 volts and less than or equal to 255 volts entries entries or Null.
Programmatic Pattern: ((RangeP[0*Volt, 255*Volt] | {(RangeP[0*Volt, 255*Volt] | Null)..} | {{RangeP[0*Volt, 255*Volt]..}..}) | Automatic) | Null
Index Matches to: Standard
StandardInclusionDeclusteringVoltage
The overriding source voltage value that can be applied to the StandardInclusionMass. Null will default to the StandardDeclusteringVoltage option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts or Null entries or list of one or more list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts entries entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 150*Volt] | {(RangeP[0.1*Volt, 150*Volt] | Null)..} | {{RangeP[0.1*Volt, 150*Volt]..}..}) | Automatic) | Null
Index Matches to: Standard
StandardInclusionChargeState
The maximum charge state of the StandardInclusionMass to also consider for inclusion. For example, if this is set to 3 and the polarity is Positive, then +1,+2,+3 charge states will be considered as well.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 or Null entries or list of one or more list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 entries entries or Null.
Programmatic Pattern: ((RangeP[0, 6, 1] | {(RangeP[0, 6, 1] | Null)..} | {{RangeP[0, 6, 1]..}..}) | Automatic) | Null
Index Matches to: Standard
StandardInclusionScanTime
The overriding scan time duration that can be applied to the StandardInclusionMass for the consequent fragmentation. Null will default to the StandardFragmentScanTime option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null entries or list of one or more list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {(RangeP[0.015*Second, 10*Second] | Null)..} | {{RangeP[0.015*Second, 10*Second]..}..}) | Automatic) | Null
Index Matches to: Standard
StandardInclusionMassTolerance
The range above and below each ion in StandardInclusionMass to consider for prioritization. For example, if set to 0.5 Gram/Mole, the total range is 1 Gram/Mole.
Default Calculation: Set to 0.5 Gram/Mole if StandardInclusionMass is given; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardSurveyChargeStateExclusion
Indicates if redundant ions that differ by ionic charge (+1/-1, +2/-2, etc.) should be excluded and if StandardChargeState exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the StandardChargeState options are set; otherwise, False.
Pattern Description: List of one or more True or False or Null entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {(BooleanP | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardSurveyIsotopeExclusion
Indicates if redundant ions that differ by isotopic mass (e.g. 1, 2 Gram/Mole) should be excluded and if StandardMassIsotope exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the StandardIsotopeExclusion options are set; otherwise, False.
Pattern Description: List of one or more True or False or Null entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {(BooleanP | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardChargeStateExclusionLimit
The number of ions to survey first with exclusion by ionic state. For example, if StandardAcquisitionSurvey is 10 and this option is 5, then 5 ions will be surveyed with charge-state exclusion. For candidate ions of rank 6 to 10, no exclusion will be performed.
Default Calculation: Inherited from any supplied method; otherwise, set the same to StandardAcquisitionSurvey, if any ChargeState option is set.
Pattern Description: Greater than or equal to 0 and less than or equal to 30 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 30 in increments of 1 or Null entries or Null.
Programmatic Pattern: ((RangeP[0, 30, 1] | {(RangeP[0, 30, 1] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardChargeStateExclusion
The specific ionic states of intact ions to redundantly exclude from the survey for further fragmentation/acquisition. 1 refers to +1/-1, 2 refers to +2/-2, etc.
Default Calculation: When StandardSurveyChargeStateExclusion is True, set to {1,2}; otherwise, Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 entries or Null entries or Null.
Programmatic Pattern: ((RangeP[1, 6, 1] | {(RangeP[1, 6, 1] | Null | {RangeP[1, 6, 1]..})..}) | Automatic) | Null
Index Matches to: Standard
StandardChargeStateMassTolerance
The range of m/z to consider for exclusion by ionic state property when StandardSurveyChargeStateExclusion is True.
Default Calculation: When StandardSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardIsotopicExclusion
The m/z difference between monoisotopic ions as a criterion for survey exclusion.
Default Calculation: When StandardSurveyIsotopeExclusion is True, set to 1 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole entries or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null | {RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]..})..}) | Automatic) | Null
Index Matches to: Standard
StandardIsotopeRatioThreshold
The minimum relative magnitude between monoisotopic ions in order to be considered for exclusion.
Default Calculation: When StandardSurveyIsotopeExclusion is True, set to 0.1; otherwise, Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 entries or Null entries or Null.
Programmatic Pattern: ((RangeP[0, 1] | {(RangeP[0, 1] | Null | {RangeP[0, 1]..})..}) | Automatic) | Null
Index Matches to: Standard
StandardIsotopeDetectionMinimum
The acquisition rate of a given intact mass to consider for isotope exclusion in the survey.
Default Calculation: When StandardSurveyIsotopeExclusion is True, set to 10 1/Second; otherwise, Null.
Pattern Description: Greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds entries or Null entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(1/Second)] | {(GreaterEqualP[0*(1/Second)] | Null | {GreaterEqualP[0*(1/Second)]..})..}) | Automatic) | Null
Index Matches to: Standard
StandardIsotopeMassTolerance
The range of m/z around a mass to consider for exclusion. This applies for both ChargeState and mass shifted Isotope. If set to 0.5 Gram/Mole, then the total range should be 1 Gram/Mole.
Default Calculation: When StandardSurveyIsotopeExclusion or StandardSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardIsotopeRatioTolerance
The range of relative magnitude around StandardIsotopeRatio to consider for isotope exclusion.
Default Calculation: If StandardSurveyIsotopeExclusion is True, set to 30*Percent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more greater than or equal to 0 percent and less than or equal to 100 percent or Null entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {(RangeP[0*Percent, 100*Percent] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardDwellTime
The duration of time for which spectra are acquired at the specific mass detection value for SelectedIonMonitoring and MultipleReactionMonitoring mode in ESI-QQQ.
Default Calculation: Is automatically set to 200 microsecond if StandardAcquisition is in SelectedIonMonitoring or MultipleReactionMonitoring mode. Otherwise is set to Null.
Pattern Description: Greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds entries or Null entries or Null.
Programmatic Pattern: ((RangeP[5*Millisecond, 2000*Millisecond] | {(RangeP[5*Millisecond, 2000*Millisecond] | Null | {RangeP[5*Millisecond, 2000*Millisecond]..})..}) | Automatic) | Null
Index Matches to: Standard
StandardCollisionCellExitVoltage
Also known as the Collision Cell Exit Potential (CXP). This value focuses and accelerates the ions out of collision cell (Q2) and into 2nd mass analyzer (MS 2). This potential is tuned to ensure successful ion acceleration out of collision cell and into MS2, and can be adjusted to reach the maximal signal intensity. This option is unique to ESI-QQQ for now, and only required when Fragment ->True and/or in ScanMode that achieves tandem mass feature (PrecursorIonScan, NeutralIonLoss,ProductIonScan,MultipleReactionMonitoring). For non-tandem mass ScanMode (FullScan and SelectedIonMonitoring) and other massspectrometer (ESI-QTOF and MALDI-TOF), this option is resolved to Null.
Default Calculation: For TripleQuadrupole as the MassAnalyzer, is set to first CollisionCellExitVoltage, otherwise set to Null.
Pattern Description: Greater than or equal to -55 volts and less than or equal to 55 volts or list of one or more greater than or equal to -55 volts and less than or equal to 55 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[-55*Volt, 55*Volt] | {(RangeP[-55*Volt, 55*Volt] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardMassDetectionStepSize
Indicate the step size for mass collection in range when using TripleQuadruploe as the MassAnalyzer.
Default Calculation: Is set to first MassDetectionStepSize
Pattern Description: Greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or list of one or more greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | {(RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardNeutralLoss
A neutral loss scan is performed on ESI-QQQ mass spectrometry by scanning the sample through the first quadrupole (Q1). The ions are then fragmented in the collision cell. The second mass analyzer is then scanned with a fixed offset to MS1. This option represents the value of this offset.
Default Calculation: Is set to 500 g/mol if using NeutralIonLoss as StandardAcquisitionMode, and is Null in other modes.
Pattern Description: Greater than 0 grams per mole or list of one or more greater than 0 grams per mole or Null entries or Null.
Programmatic Pattern: ((GreaterP[0*(Gram/Mole)] | {(GreaterP[0*(Gram/Mole)] | Null)..}) | Automatic) | Null
Index Matches to: Standard
StandardMultipleReactionMonitoringAssays
In ESI-QQQ mass spectrometry analysis, the ion corresponding to the compound of interest is targeted with subsequent fragmentation of that target ion to produce a range of daughter ions. One (or more) of these fragment daughter ions can be selected for quantitation purposes. Only compounds that meet both these criteria, i.e. specific parent ion and specific daughter ions corresponding to the mass of the molecule of interest are detected within the mass spectrometer. The mass assays (MS1/MS2 mass value combinations) for each scan, along with the CollisionEnergy and DwellTime (length of time of each scan).
Default Calculation: Is set based on StandardMassDetection, StandardCollisionEnergy, StandardDwellTime and StandardFragmentMassDetection.
Pattern Description: List of one or more Individual Multiple Reaction Monitoring Assay or {Multiple Reaction Monitoring Assays} or None entries or Null.
Programmatic Pattern: (({({{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..} | {{{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..}} | (Null | {Null}))..} | Null) | Automatic) | Null
Index Matches to: Standard
StandardAbsorbanceWavelength
For Standard measurement, the wavelength of light passed through the flow cell for the PhotoDiodeArray (PDA) Detector. A 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 500*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 490*Nanometer, 1*Nanometer] ;; RangeP[200*Nanometer, 500*Nanometer, 1*Nanometer]) | Automatic) | Null
Index Matches to: Standard
StandardWavelengthResolution
The increment in wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector for Standard measurement.
Default Calculation: Automatically set to the same as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[1.2*Nanometer, 12.*Nanometer] | Automatic) | Null
Index Matches to: Standard
StandardUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the sample for the PhotoDiodeArray (PDA) detector for Standard measurement.
Default Calculation: Automatically set to the same as the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: Standard
StandardAbsorbanceSamplingRate
The number of times an absorbance measurement is made per second for Standard sample. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second), 1*(1/Second)] | Automatic) | Null
Index Matches to: Standard
StandardStorageCondition
The non-default conditions under which any standards used by this experiment should be stored after the protocol is completed. If left unset, the standard samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Index Matches to: Standard
Blanks
Blank
The object(s) (samples) to inject typically as negative controls (e.g. to test effects stemming from injection, sample solvent, impurities on the column or buffer).
Default Calculation: If any other Blank option is specified or RefractiveIndex Detector is selected, automatically set to the specified BufferA or Model[Sample, "Milli-Q water"].
Pattern Description: An object of type or subtype Model[Sample] or Object[Sample] or a prepared sample or Null.
Programmatic Pattern: ((ObjectP[{Model[Sample], Object[Sample]}] | _String) | Automatic) | Null
BlankInjectionVolume
The physical quantity of Blank sample that is loaded into the flow path on the selected instrument along with the mobile phase onto the stationary phase.
Default Calculation: Automatically set equal to the first entry in InjectionVolume.
Pattern Description: Greater than or equal to 0 microliters and less than or equal to 500 microliters or Null.
Programmatic Pattern: (RangeP[0*Microliter, 500*Microliter] | Automatic) | Null
BlankFrequency
The frequency at which Blank measurements will be inserted between Sample.
Default Calculation: If injectionTable is given, automatically set to Null and the sequence of Blanks specified in the InjectionTable will be used in the experiment. If any other Blank option is specified, automatically set to FirstAndLast.
Pattern Description: Greater than 0 in increments of 1 or None, First, Last, FirstAndLast, or GradientChange or Null.
Programmatic Pattern: (((None | First | Last | FirstAndLast | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
BlankColumnTemperature
The temperature of the column when the Blank gradient and measurement are run. If BlankColumnTemperature is set to Ambient, column oven temperature control is not used. Otherwise, BlankColumnTemperature is maintained by temperature control of the column oven.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the BlankGradient option or the column temperature for the sample in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 30 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[30*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
BlankGradientA
The composition of BufferA within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientB, BlankGradientC, and BlankGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientB
The composition of BufferB within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientA, BlankGradientC, and BlankGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientC
The composition of BufferC within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientA, BlankGradientB, and BlankGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankGradientD
The composition of BufferD within the flow, defined for specific time points for Blank measurement. The composition is linearly interpolated for the intervening periods between the defined time points. For example for BlankGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If BlankGradient option is specified, set from it or implicitly determined from the BlankGradientA, BlankGradientB, and BlankGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
BlankFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the BlankGradient options during Blank measurement. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If BlankGradient option is specified, automatically set from the method given in the BlankGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 2 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
BlankGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow during Blank measurement. Specific parameters of a gradient object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the BlankGradient options (e.g. BlankGradientA, BlankGradientB, BlankGradientC, BlankGradientD, BlankFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
BlankAnalytes
The compounds of interest that are present in the given Blank samples, used to determine the other settings for the Mass Spectrometer (e.g. MassRange).
Default Calculation: If populated, will resolve to the user-specified Analytes field in the Object[Sample]. Otherwise, will resolve to the larger compounds in the sample, in the order of Proteins, Peptides, Oligomers, then other small molecules. Otherwise, set Null.
Pattern Description: List of one or more an object of type or subtype Model[Molecule], Model[Molecule, cDNA], Model[Molecule, Oligomer], Model[Molecule, Transcript], Model[Molecule, Protein], Model[Molecule, Protein, Antibody], Model[Molecule, Carbohydrate], Model[Molecule, Polymer], Model[Resin], Model[Resin, SolidPhaseSupport], Model[Lysate], Model[ProprietaryFormulation], Model[Virus], Model[Cell], Model[Cell, Mammalian], Model[Cell, Bacteria], Model[Cell, Yeast], Model[Tissue], Model[Material], or Model[Species] entries or Null.
Programmatic Pattern: ({ObjectP[IdentityModelTypes]..} | Automatic) | Null
BlankIonMode
Indicates if positively or negatively charged ions are analyzed.
Default Calculation: Set to the first IonMode for an analyte input sample.
Pattern Description: Negative or Positive or Null.
Programmatic Pattern: (IonModeP | Automatic) | Null
BlankMassSpectrometryMethod
The previously specified instruction(s) for the analyte ionization, selection, fragmentation, and detection.
Default Calculation: If Blank samples exist and MassSpectrometryMethod is specified, then set to the first available BlankMassSpectrometryMethod.
Pattern Description: An object of type or subtype Object[Method, MassAcquisition] or New or Null.
Programmatic Pattern: ((ObjectP[Object[Method, MassAcquisition]] | New) | Automatic) | Null
BlankESICapillaryVoltage
The absolute voltage applied to the tip of the stainless steel capillary tubing in order to produce charged droplets. Adjust this voltage to maximize sensitivity. Most compounds are optimized between 0.5 and 3.2 kV in ESI positive ion mode and 0.5 and 2.6 in ESI negative ion mode, but can be altered according to sample type. For low flow applications, best sensitivity will be achieved with a relatively high value in ESI positive (e.g. 3.0 kV), for blank flow UPLC a value of 0.5 kV is typically best for maximum sensitivity.
Default Calculation: Is automatically set to the first ESICapillaryVoltage.
Pattern Description: Greater than or equal to -4 kilovolts and less than or equal to 5 kilovolts or Null.
Programmatic Pattern: (RangeP[-4*Kilovolt, 5*Kilovolt] | Automatic) | Null
BlankDeclusteringVoltage
The voltage offset between the ion block (the reduced pressure chamber of the source block) and the stepwave ion guide (the optics before the quadrupole mass analyzer). This voltage attracts charged ions in the spray being produced from the capillary tip into the ion block leading into the mass spectrometer. This voltage is typically set to 25-100 V and its tuning has little effect on sensitivity compared to other options (e.g. BlankStepwaveVoltage).
Default Calculation: Is automatically set to any specified MassAcquisition method; otherwise, set to 40 Volt.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 150*Volt] | Automatic) | Null
BlankStepwaveVoltage
The voltage offset between the 1st and 2nd stage of the stepwave ion guide which leads ions coming from the sample cone towards the quadrupole mass analyzer. This voltage normally optimizes between 25 and 150 V and should be adjusted for sensitivity depending on compound and charge state. For multiply charged species it is typically set to to 40-50 V, and higher for singly charged species. In general higher cone voltages (120-150 V) are needed for larger mass ions such as intact proteins and monoclonal antibodies. It also has greatest effect on in-source fragmentation and should be decreased if in-source fragmentation is observed but not desired.
Default Calculation: Is automatically set to the first StepwaveVoltage.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 200 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 200*Volt] | Automatic) | Null
BlankSourceTemperature
The temperature setting of the source block. Heating the source block discourages condensation and decreases solvent clustering in the reduced vacuum region of the source. This temperature setting is flow rate and sample dependent. Typical values are between 60 to 120 Celsius. For thermally labile analytes, a lower temperature setting is recommended.
Default Calculation: Is automatically set to the first SourceTemperature.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 150*Celsius] | Automatic) | Null
BlankDesolvationTemperature
The temperature setting for the ESI desolvation heater that controls the nitrogen gas temperature used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to BlankDesolvationGasFlow, this setting is dependent on solvent flow rate and composition. A typical range is from 150 to 650 Celsius.
Default Calculation: Is automatically set to the first DesolvationTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 650 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 650*Celsius] | Automatic) | Null
BlankDesolvationGasFlow
The rate at which nitrogen gas is flowed around the ESI capillary. It is used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to BlankDesolvationTemperature, this setting is dependent on solvent flow rate and composition. Higher desolvation temperatures usually result in increased sensitivity, but too high values can cause spray instability. Typical values are between 300 to 1200 L/h.
Default Calculation: Is automatically set to the first DesolvationGasFlow.
Pattern Description: Greater than or equal to 55 liters per hour and less than or equal to 1200 liters per hour or greater than or equal to 0 pounds
‐force per inch squared and less than or equal to 85 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[55*(Liter/Hour), 1200*(Liter/Hour)] | RangeP[0*PSI, 85*PSI]) | Automatic) | Null
BlankConeGasFlow
The rate at which nitrogen gas is flowed around the sample inlet cone (the spherical metal plate acting as a first gate between the sprayer and the reduced pressure chamber, the ion block). This gas flow is used to minimize the formation of solvent ion clusters. It also helps reduce adduct ions and directing the spray into the ion block while keeping the sample cone clean. Typical values are between 0 and 150 L/h.
Default Calculation: Is automatically set to the first ConeGasFlow.
Pattern Description: Greater than or equal to 0 liters per hour and less than or equal to 300 liters per hour or greater than or equal to 30 pounds
‐force per inch squared and less than or equal to 55 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[0*(Liter/Hour), 300*(Liter/Hour)] | RangeP[30*PSI, 55*PSI]) | Automatic) | Null
BlankAcquisitionWindow
The time range with respect to the chromatographic separation to conduct analyte ionization, selection/survey, optional fragmentation, and detection.
Default Calculation: Set to the entire gradient window 0 Minute to the last time point in BlankGradient.
Pattern Description: A span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours or list of one or more a span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours entries or Null.
Programmatic Pattern: ((RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour] | {(Alternatives[RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour]])..}) | Automatic) | Null
BlankAcquisitionMode
The method by which spectra are collected. DataDependent will depend on the properties of the measured mass spectrum of the intact ions. DataIndependent will systemically scan through all of the intact ions. MS1 will focus on defined intact masses. MS1MS2 will focus on fragmented masses.
Default Calculation: Set to MS1FullScan unless DataDependent related options are set, then set to DataDependent.
Pattern Description: DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring or list of one or more DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring entries or Null.
Programmatic Pattern: ((MSAcquisitionModeP | {MSAcquisitionModeP..}) | Automatic) | Null
BlankFragment
Indicates if ions should be collided with neutral gas and dissociated in order to measure the resulting product ions. Also known as tandem mass spectrometry or MS/MS (as opposed to MS).
Default Calculation: Set to True if BlankAcquisitionMode is MS1MS2ProductIonScan, DataDependent, or DataIndependent. Set True if any of the Fragmentation related options are set (e.g. BlankFragmentMassDetection).
Pattern Description: List of one or more True or False entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {BooleanP..}) | Automatic) | Null
BlankMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for intact masses. When BlankFragment is True, the intact ions will be selected for fragmentation.
Default Calculation: For BlankFragment -> False, automatically set to one of three default mass ranges according to the molecular weight of the BlankAnalytes to encompass them.
Pattern Description: All or Range or Single or Specific List or list of one or more All or Range or Single or Specific List entries or Null.
Programmatic Pattern: (((RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | MSAnalyteGroupP) | {(RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | MSAnalyteGroupP)..}) | Automatic) | Null
BlankScanTime
The duration of time allowed to pass between each spectral acquisition. When BlankAcquisitionMode is DataDependent, this value refers to the duration for measuring spectra from the intact ions. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired.
Default Calculation: Set to 0.2 seconds unless a method is given.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {RangeP[0.015*Second, 10*Second]..}) | Automatic) | Null
BlankFragmentMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for product ions. When BlankAcquisitionMode is DataDependent|DataIndependent, all of the product ions in consideration for measurement. Null if BlankFragment is False.
Default Calculation: When BlankFragment is False, set to Null. Otherwise, 20 Gram/Mole to the maximum BlankMassDetection.
Pattern Description: DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null or list of one or more DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null entries.
Programmatic Pattern: ((({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..}) | {({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..})..}) | Automatic) | Null
BlankCollisionEnergy
The voltage by which intact ions are accelerated through inert gas in order to dissociate them into measurable fragment ion species when BlankFragment is True. BlankCollisionEnergy cannot be defined simultaneously with BlankCollisionEnergyMassProfile.
Default Calculation: Is automatically set to 40 Volt when BlankFragment is True, otherwise is set to Null.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 255 volts or greater than or equal to -180 volts and less than or equal to 5 volts or list of one or more A list of Single Values or Single Values or Null entries or Null.
Programmatic Pattern: (((RangeP[0.1*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {((RangeP[0.1*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {(RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt])..} | Null)..}) | Automatic) | Null
BlankCollisionEnergyMassProfile
The relationship of collision energy with the BlankMassDetection.
Default Calculation: Set to BlankCollisionEnergyMassScan if defined; otherwise, set to Null.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or list of one or more a span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | {(RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Null)..}) | Automatic) | Null
BlankCollisionEnergyMassScan
The collision energy profile at the end of the scan from BlankCollisionEnergy or BlankCollisionEnergyScanProfile, as related to analyte mass.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or list of one or more a span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | {(RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Null)..}) | Automatic) | Null
BlankFragmentScanTime
The duration of the spectral scanning for each fragmentation of an intact ion when BlankAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to the same value as ScanTime if BlankAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {(RangeP[0.015*Second, 10*Second] | Null)..}) | Automatic) | Null
BlankAcquisitionSurvey
The number of intact ions to consider for fragmentation and product ion measurement in every measurement cycle when BlankAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to 10 if BlankAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 30 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 30 in increments of 1 or Null entries or Null.
Programmatic Pattern: ((RangeP[1, 30, 1] | {(RangeP[1, 30, 1] | Null)..}) | Automatic) | Null
BlankMinimumThreshold
The minimum number of total ions detected within ScanTime durations needed to trigger the start of data dependent acquisition when BlankAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to (100000/Second)*ScanTime if BlankAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or list of one or more greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null entries or Null.
Programmatic Pattern: ((RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | {(RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Null)..}) | Automatic) | Null
BlankAcquisitionLimit
The maximum number of total ions for a specific intact ion when BlankAcquisitionMode is set to DataDependent. When this value is exceeded, acquisition will switch to fragmentation of the next candidate ion.
Default Calculation: Automatically inherited from supplied method if BlankAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or list of one or more greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null entries or Null.
Programmatic Pattern: ((RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | {(RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Null)..}) | Automatic) | Null
BlankCycleTimeLimit
The maximum possible computed duration of all of the scans for the intact and fragmentation measurements when BlankAcquisitionMode is set to DataDependent.
Default Calculation: Calculated from the BlankAcquisitionSurvey, BlankScanTime, and BlankFragmentScanTime.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 20000*Second] | {(RangeP[0.015*Second, 20000*Second] | Null)..}) | Automatic) | Null
BlankExclusionDomain
The time range when the BlankExclusionMasses are omitted in the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire BlankAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or Null entries or list of one or more list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full | Null)..} | {{(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}..}) | Automatic) | Null
BlankExclusionMass
The intact ions (Target Mass) to omit. When the Mode is set to All, the mass is excluded for the entire ExclusionDomain. When the Mode is set to Once, the Mass is excluded in the first survey appearance, but considered for consequent ones.
Default Calculation: If any BlankExclusionMode-related options are set (e.g. BlankExclusionMassTolerance), a target mass of the first Analyte (if not in BlankInclusionMasses) is chosen and retention time is set to 0*Minute.
Pattern Description: List of one or more list of one or more {Mode, Target Mass} entries entries or list of one or more {Mode, Target Mass} or Null entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | {({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | Null)..} | {{{All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]}..}..}) | Automatic) | Null
BlankExclusionMassTolerance
The range above and below each ion in BlankExclusionMasses to consider for omission when BlankExclusionMass is All or Once.
Default Calculation: If BlankExclusionMass -> All or Once, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
BlankExclusionRetentionTimeTolerance
The range of time above and below the BlankExclusionDomain to consider for exclusion.
Default Calculation: If BlankExclusionMass and BlankExclusionDomain options are set, this is set to 10 seconds; otherwise, Null.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 3600 seconds or list of one or more greater than or equal to 0 seconds and less than or equal to 3600 seconds or Null entries or Null.
Programmatic Pattern: ((RangeP[0*Second, 3600*Second] | {(RangeP[0*Second, 3600*Second] | Null)..}) | Automatic) | Null
BlankInclusionDomain
The time range when BlankInclusionMass applies with respect to the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire BlankAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or Null entries or list of one or more list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full | Null)..} | {{(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}..}) | Automatic) | Null
BlankInclusionMass
The ions (Target Mass) to prioritize during the survey scan for further fragmentation when BlankAcquisitionMode is DataDependent. BlankInclusionMass set to Only will solely be considered for surveys. When Mode is Preferential, the InclusionMass will be prioritized for survey.
Default Calculation: When BlankInclusionMode Only or Preferential, an entry mass is added based on the mass of the most salient analyte of the sample.
Pattern Description: List of one or more list of one or more {Mode, Target Mass} entries entries or list of one or more {Mode, Target Mass} or Null entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | {({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | Null)..} | {{{Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]}..}..}) | Automatic) | Null
BlankInclusionCollisionEnergy
The overriding collision energy value that can be applied to the BlankInclusionMass. Null will default to the BlankCollisionEnergy option and related.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 volts and less than or equal to 255 volts or list of one or more greater than or equal to 0 volts and less than or equal to 255 volts or Null entries or list of one or more list of one or more greater than or equal to 0 volts and less than or equal to 255 volts entries entries or Null.
Programmatic Pattern: ((RangeP[0*Volt, 255*Volt] | {(RangeP[0*Volt, 255*Volt] | Null)..} | {{RangeP[0*Volt, 255*Volt]..}..}) | Automatic) | Null
BlankInclusionDeclusteringVoltage
The overriding source voltage value that can be applied to the BlankInclusionMass. Null will default to the BlankDeclusteringVoltage option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts or Null entries or list of one or more list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts entries entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 150*Volt] | {(RangeP[0.1*Volt, 150*Volt] | Null)..} | {{RangeP[0.1*Volt, 150*Volt]..}..}) | Automatic) | Null
BlankInclusionChargeState
The maximum charge state of the BlankInclusionMass to also consider for inclusion. For example, if this is set to 3 and the polarity is Positive, then +1,+2,+3 charge states will be considered as well.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 or Null entries or list of one or more list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 entries entries or Null.
Programmatic Pattern: ((RangeP[0, 6, 1] | {(RangeP[0, 6, 1] | Null)..} | {{RangeP[0, 6, 1]..}..}) | Automatic) | Null
BlankInclusionScanTime
The overriding scan time duration that can be applied to the BlankInclusionMass for the consequent fragmentation. Null will default to the BlankFragmentScanTime option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null entries or list of one or more list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {(RangeP[0.015*Second, 10*Second] | Null)..} | {{RangeP[0.015*Second, 10*Second]..}..}) | Automatic) | Null
BlankInclusionMassTolerance
The range above and below each ion in BlankInclusionMass to consider for prioritization. For example, if set to 0.5 Gram/Mole, the total range is 1 Gram/Mole.
Default Calculation: Set to 0.5 Gram/Mole if BlankInclusionMass is given; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
BlankSurveyChargeStateExclusion
Indicates if redundant ions that differ by ionic charge (+1/-1, +2/-2, etc.) should be left out and if BlankChargeState exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the BlankChargeState options are set; otherwise, False.
Pattern Description: List of one or more True or False or Null entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {(BooleanP | Null)..}) | Automatic) | Null
BlankSurveyIsotopeExclusion
Indicates if redundant ions that differ by isotopic mass (e.g. 1, 2 Gram/Mole) should be excluded and if BlankMassIsotope exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the BlankIsotopeExclusion options are set; otherwise, False.
Pattern Description: List of one or more True or False or Null entries or True or False or Null.
Programmatic Pattern: ((BooleanP | {(BooleanP | Null)..}) | Automatic) | Null
BlankChargeStateExclusionLimit
The number of ions to survey first with exclusion by ionic state. For example, if BlankAcquisitionSurvey is 10 and this option is 5, then 5 ions will be surveyed with charge-state exclusion. For candidate ions of rank 6 to 10, no exclusion will be performed.
Default Calculation: Inherited from any supplied method; otherwise, set the same to BlankAcquisitionSurvey, if any ChargeState option is set.
Pattern Description: Greater than or equal to 0 and less than or equal to 30 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 30 in increments of 1 or Null entries or Null.
Programmatic Pattern: ((RangeP[0, 30, 1] | {(RangeP[0, 30, 1] | Null)..}) | Automatic) | Null
BlankChargeStateExclusion
The specific ionic states of intact ions to redundantly exclude from the survey for further fragmentation/acquisition. 1 refers to +1/-1, 2 refers to +2/-2, etc.
Default Calculation: When BlankSurveyChargeStateExclusion is True, set to {1,2}; otherwise, Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 entries or Null entries or Null.
Programmatic Pattern: ((RangeP[1, 6, 1] | {(RangeP[1, 6, 1] | Null | {RangeP[1, 6, 1]..})..}) | Automatic) | Null
BlankChargeStateMassTolerance
The range of m/z to consider for exclusion by ionic state property when BlankSurveyChargeStateExclusion is True.
Default Calculation: When BlankSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
BlankIsotopicExclusion
The m/z difference between monoisotopic ions as a criterion for survey exclusion.
Default Calculation: When BlankSurveyIsotopeExclusion is True, set to 1 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole entries or Null entries or Null.
Programmatic Pattern: (((Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]]) | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null | {RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]..})..}) | Automatic) | Null
BlankIsotopeRatioThreshold
The minimum relative magnitude between monoisotopic ions in order to be considered an isotope for exclusion.
Default Calculation: When BlankSurveyIsotopeExclusion is True, set to 0.1; otherwise, Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 entries or Null entries or Null.
Programmatic Pattern: ((RangeP[0, 1] | {(RangeP[0, 1] | Null | {RangeP[0, 1]..})..}) | Automatic) | Null
BlankIsotopeDetectionMinimum
The acquisition rate of a given intact mass to consider for isotope exclusion in the survey.
Default Calculation: When BlankSurveyIsotopeExclusion is True, set to 10 1/Second; otherwise, Null.
Pattern Description: Greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds entries or Null entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(1/Second)] | {(GreaterEqualP[0*(1/Second)] | Null | {GreaterEqualP[0*(1/Second)]..})..}) | Automatic) | Null
BlankIsotopeMassTolerance
The range of m/z around a mass to consider for exclusion. This applies for both ChargeState and mass shifted Isotope. If set to 0.5 Gram/Mole, then the total range should be 1 Gram/Mole.
Default Calculation: When BlankSurveyIsotopeExclusion or BlankSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | {(RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Null)..}) | Automatic) | Null
BlankIsotopeRatioTolerance
The range of relative magnitude around BlankIsotopeRatio to consider for isotope exclusion.
Default Calculation: If BlankSurveyIsotopeExclusion is True, set to 30*Percent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more greater than or equal to 0 percent and less than or equal to 100 percent or Null entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {(RangeP[0*Percent, 100*Percent] | Null)..}) | Automatic) | Null
BlankNeutralLoss
A neutral loss scan is performed on ESI-QQQ mass spectrometry by scanning the sample through the first quadrupole (Q1). The ions are then fragmented in the collision cell. The second mass analyzer is then scanned with a fixed offset to MS1. This option represents the value of this offset.
Default Calculation: Is set to 500 g/mol if using NeutralIonLoss as the BlankAcquisitionMode, and is Null in other modes.
Pattern Description: Greater than 0 grams per mole or list of one or more greater than 0 grams per mole or Null entries or Null.
Programmatic Pattern: ((GreaterP[0*(Gram/Mole)] | {(GreaterP[0*(Gram/Mole)] | Null)..}) | Automatic) | Null
BlankDwellTime
The duration of time for which spectra are acquired at the specific mass detection value for SelectedIonMonitoring and MultipleReactionMonitoring mode in ESI-QQQ.
Default Calculation: Is automatically set to 200 microsecond if BlankAcquisition is in SelectedIonMonitoring or MultipleReactionMonitoring mode.
Pattern Description: Greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds entries or Null entries or Null.
Programmatic Pattern: ((RangeP[5*Millisecond, 2000*Millisecond] | {(RangeP[5*Millisecond, 2000*Millisecond] | Null | {RangeP[5*Millisecond, 2000*Millisecond]..})..}) | Automatic) | Null
BlankCollisionCellExitVoltage
Also known as the Collision Cell Exit Potential (CXP). This value focuses and accelerates the ions out of collision cell (Q2) and into 2nd mass analyzer (MS 2). This potential is tuned to ensure successful ion acceleration out of collision cell and into MS2, and can be adjusted to reach the maximal signal intensity. This option is unique to ESI-QQQ for now, and only required when Fragment ->True and/or in ScanMode that achieves tandem mass feature (PrecursorIonScan, NeutralIonLoss,ProductIonScan,MultipleReactionMonitoring). For non-tandem mass ScanMode (FullScan and SelectedIonMonitoring) and other massspectrometer (ESI-QTOF and MALDI-TOF), this option is resolved to Null.
Default Calculation: For TripleQuadrupole as the MassAnalyzer, is set to first CollisionCellExitVoltage, otherwise set to Null.
Pattern Description: Greater than or equal to -55 volts and less than or equal to 55 volts or list of one or more greater than or equal to -55 volts and less than or equal to 55 volts or Null entries or Null.
Programmatic Pattern: ((RangeP[-55*Volt, 55*Volt] | {(RangeP[-55*Volt, 55*Volt] | Null)..}) | Automatic) | Null
BlankMassDetectionStepSize
Indicate the step size for mass collection in range when using TripleQuadruploe as the MassAnalyzer.
Default Calculation: This option will be set to Null if using ESI-QTOF. For ESI-QQQ, if both of the mass analyzer are in mass selection mode (SelectedIonMonitoring and MultipleReactionMonitoring mode), this option will be auto resolved to Null. In all other mass scan modes in ESI-QQQ, this option will be automatically resolved to 0.1 g/mol.
Pattern Description: Greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or list of one or more greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or Null entries or Null.
Programmatic Pattern: ((RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | {(RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | Null)..}) | Automatic) | Null
BlankMultipleReactionMonitoringAssays
In ESI-QQQ mass spectrometry analysis, the ion corresponding to the compound of interest is targeted with subsequent fragmentation of that target ion to produce a range of daughter ions. One (or more) of these fragment daughter ions can be selected for quantitation purposes. Only compounds that meet both these criteria, i.e. specific parent ion and specific daughter ions corresponding to the mass of the molecule of interest are detected within the mass spectrometer. The mass assays (MS1/MS2 mass value combinations) for each scan, along with the CollisionEnergy and DwellTime (length of time of each scan).
Default Calculation: Is set based on BlankMassDetection, BlankCollisionEnergy, BlankDwellTime and BlankFragmentMassDetection.
Pattern Description: List of one or more Individual Multiple Reaction Monitoring Assay or {Multiple Reaction Monitoring Assays} or None entries or Null.
Programmatic Pattern: (({({{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..} | {{{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..}} | (Null | {Null}))..} | Null) | Automatic) | Null
BlankAbsorbanceWavelength
For Blank measurement, the wavelength of light passed through the flow cell for the UVVis Detector. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 500*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 490*Nanometer, 1*Nanometer] ;; RangeP[200*Nanometer, 500*Nanometer, 1*Nanometer]) | Automatic) | Null
BlankWavelengthResolution
The increment in wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector for Blank measurement.
Default Calculation: Automatically set to the same as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[1.2*Nanometer, 12.*Nanometer] | Automatic) | Null
BlankUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the sample for the PhotoDiodeArray Detector for Blank measurement.
Default Calculation: Automatically set to the same as the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
BlankAbsorbanceSamplingRate
The number of times the absorbance measurement is made per second during Blank measurement. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second), 1*(1/Second)] | Automatic) | Null
BlankStorageCondition
The non-default conditions under which any blanks used by this experiment should be stored after the protocol is completed. If left unset, Blank samples will be stored according to their Models' DefaultStorageCondition.
Pattern Description: {AmbientStorage, EnclosedAmbientStorage, Refrigerator, Freezer, DeepFreezer, CryogenicStorage, YeastIncubation, YeastShakingIncubation, BacterialIncubation, BacterialShakingIncubation, MammalianIncubation, ViralIncubation, CrystalIncubation, AcceleratedTesting, IntermediateTesting, LongTermTesting, UVVisLightTesting} or Disposal or Null.
Programmatic Pattern: (Alternatives[SampleStorageTypeP | Disposal]) | Null
Column Prime
ColumnRefreshFrequency
The frequency of column prime inserted into the order of analyte injections at which solvent is flowed to equilibrate the column in order to remove contaminants and reset the gradient to match the starting percentage of the subsequent injection. An initial column prime and final column flush will be performed unless Null or None is specified. For First, it is performed at the beginning. For Last, it is performed at the end. For FirstAndLast, it is performed both at the beginning and end. For GradientChange, it is performed every time a change in the gradient is encountered for the injections. A Number indicates the number of injections after which it is performed and also in the beginning (eg: for 2, it is performed at the start and after 2nd, 4th, 6th and so on injections).
Default Calculation: Automatically set to Null when InjectionTable option is specified (meaning that this option is inconsequential) or no column is used in the experiment; otherwise, set to GradientChange.
Pattern Description: Greater than 0 in increments of 1 or None, FirstAndLast, First, Last, or GradientChange or Null.
Programmatic Pattern: (((None | FirstAndLast | First | Last | GradientChange) | GreaterP[0, 1]) | Automatic) | Null
ColumnPrimeTemperature
The column's temperature at which the column prime gradient is run. If ColumnPrimeTemperature is set to Ambient, column oven temperature control is not used. Otherwise, ColumnPrimeTemperature is maintained by temperature control of the column oven.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the ColumnPrimeGradient option or the column temperature for the column prime in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 30 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[30*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
ColumnPrimeGradientA
The composition of BufferA within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientB, ColumnPrimeGradientC, and ColumnPrimeGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeGradientB
The composition of BufferB within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientA, ColumnPrimeGradientC, and ColumnPrimeGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeGradientC
The composition of BufferC within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientA, ColumnPrimeGradientB, and ColumnPrimeGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeGradientD
The composition of BufferD within the flow, defined for specific time points for column prime. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnPrimeGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnPrimeGradient option is specified, set from it or implicitly determined from the ColumnPrimeGradientA, ColumnPrimeGradientB, and ColumnPrimeGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnPrimeFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the ColumnPrimeGradient options during column prime. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If ColumnPrimeGradient option is specified, automatically set from the method given in the ColumnPrimeGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 2 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnPrimeGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow for column prime. Specific parameters of a gradient object can be overridden by specific options.
Default Calculation: Automatically set to best meet all the ColumnPrimeGradient options (e.g. ColumnPrimeGradientA, ColumnPrimeGradientB, ColumnPrimeGradientC, ColumnPrimeGradientD, ColumnPrimeFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnPrimeIonMode
Indicates if positively or negatively charged ions are analyzed.
Default Calculation: Set to the first IonMode for an analyte input sample.
Pattern Description: Negative or Positive or Null.
Programmatic Pattern: (IonModeP | Automatic) | Null
ColumnPrimeMassSpectrometryMethod
The previously specified instruction(s) for the analyte ionization, selection, fragmentation, and detection.
Default Calculation: If ColumnPrime samples exist and MassSpectrometryMethod is specified, then set to the first available ColumnPrimeMassSpectrometryMethod.
Pattern Description: An object of type or subtype Object[Method, MassAcquisition] or New or Null.
Programmatic Pattern: ((ObjectP[Object[Method, MassAcquisition]] | New) | Automatic) | Null
ColumnPrimeESICapillaryVoltage
The absolute voltage applied to the tip of the stainless steel capillary tubing in order to produce charged droplets. Adjust this voltage to maximize sensitivity. Most compounds are optimized between 0.5 and 3.2 kV in ESI positive ion mode and 0.5 and 2.6 in ESI negative ion mode, but can be altered according to sample type. For low flow applications, best sensitivity will be achieved with a relatively high value in ESI positive (e.g. 3.0 kV), for columnPrime flow UPLC a value of 0.5 kV is typically best for maximum sensitivity.
Default Calculation: Is automatically set to the first ESICapillaryVoltage.
Pattern Description: Greater than or equal to -4 kilovolts and less than or equal to 5 kilovolts or Null.
Programmatic Pattern: (RangeP[-4*Kilovolt, 5*Kilovolt] | Automatic) | Null
ColumnPrimeDeclusteringVoltage
The voltage offset between the ion block (the reduced pressure chamber of the source block) and the stepwave ion guide (the optics before the quadrupole mass analyzer). This voltage attracts charged ions in the spray being produced from the capillary tip into the ion block leading into the mass spectrometer. This voltage is typically set to 25-100 V and its tuning has little effect on sensitivity compared to other options (e.g. ColumnPrimeStepwaveVoltage).
Default Calculation: Is automatically set to any specified MassAcquisition method; otherwise, set to 40 Volt.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 150*Volt] | Automatic) | Null
ColumnPrimeStepwaveVoltage
The voltage offset between the 1st and 2nd stage of the stepwave ion guide which leads ions coming from the sample cone towards the quadrupole mass analyzer. This voltage normally optimizes between 25 and 150 V and should be adjusted for sensitivity depending on compound and charge state. For multiply charged species it is typically set to to 40-50 V, and higher for singly charged species. In general higher cone voltages (120-150 V) are needed for larger mass ions such as intact proteins and monoclonal antibodies. It also has greatest effect on in-source fragmentation and should be decreased if in-source fragmentation is observed but not desired.
Default Calculation: Is automatically set to the first StepwaveVoltage.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 200 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 200*Volt] | Automatic) | Null
ColumnPrimeSourceTemperature
The temperature setting of the source block. Heating the source block discourages condensation and decreases solvent clustering in the reduced vacuum region of the source. This temperature setting is flow rate and sample dependent. Typical values are between 60 to 120 Celsius. For thermally labile analytes, a lower temperature setting is recommended.
Default Calculation: Is automatically set to the first SourceTemperature.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 150*Celsius] | Automatic) | Null
ColumnPrimeDesolvationTemperature
The temperature setting for the ESI desolvation heater that controls the nitrogen gas temperature used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to ColumnPrimeDesolvationGasFlow, this setting is dependent on solvent flow rate and composition. A typical range is from 150 to 650 Celsius.
Default Calculation: Is automatically set to the first DesolvationTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 650 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 650*Celsius] | Automatic) | Null
ColumnPrimeDesolvationGasFlow
The rate at which nitrogen gas is flowed around the ESI capillary. It is used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to ColumnPrimeDesolvationTemperature, this setting is dependent on solvent flow rate and composition. Higher desolvation temperatures usually result in increased sensitivity, but too high values can cause spray instability. Typical values are between 300 to 1200 L/h.
Default Calculation: Is automatically set to the first DesolvationGasFlow.
Pattern Description: Greater than or equal to 55 liters per hour and less than or equal to 1200 liters per hour or greater than or equal to 0 pounds
‐force per inch squared and less than or equal to 85 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[55*(Liter/Hour), 1200*(Liter/Hour)] | RangeP[0*PSI, 85*PSI]) | Automatic) | Null
ColumnPrimeConeGasFlow
The rate at which nitrogen gas flow is flowed around the sample inlet cone (the spherical metal plate acting as a first gate between the sprayer and the reduced pressure chamber, the ion block). This gas flow is used to minimize the formation of solvent ion clusters. It also helps reduce adduct ions and directing the spray into the ion block while keeping the sample cone clean. Typical values are between 0 and 150 L/h.
Default Calculation: Is automatically set to the first ConeGasFlow.
Pattern Description: Greater than or equal to 0 liters per hour and less than or equal to 300 liters per hour or greater than or equal to 30 pounds
‐force per inch squared and less than or equal to 55 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[0*(Liter/Hour), 300*(Liter/Hour)] | RangeP[30*PSI, 55*PSI]) | Automatic) | Null
ColumnPrimeAcquisitionWindow
The time range with respect to the chromatographic separation to conduct analyte ionization, selection/survey, optional fragmentation, and detection.
Default Calculation: Set to the entire gradient window 0 Minute to the last time point in ColumnPrimeGradient.
Pattern Description: A span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeAcquisitionMode
The method by which spectra are collected. DataDependent will depend on the properties of the measured mass spectrum of the intact ions. DataIndependent will systemically scan through all of the intact ions. MS1FullScan will focus on defined intact masses. MS1MS2 will focus on fragmented masses.
Default Calculation: Set to MS1FullScan unless DataDependent related options are set, then set to DataDependent.
Pattern Description: DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring or Null.
Programmatic Pattern: (MSAcquisitionModeP | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeFragment
Indicates if ions should be collided with neutral gas in order to measure the resulting product ions. Also known as tandem mass spectrometry or MS/MS (as opposed to MS).
Default Calculation: Set to True if ColumnPrimeAcquisitionMode is MS1MS2ProductIonScan, DataDependent, or DataIndependent. Set True if any of the Fragmentation related options are set (e.g. ColumnPrimeFragmentMassDetection).
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for intact masses. When ColumnPrimeFragment is True, the intact ions will be selected for fragmentation.
Default Calculation: For ColumnPrimeFragment -> False, automatically set to one of three default mass ranges according to the molecular weight of the ColumnPrimeAnalytes to encompass them.
Pattern Description: All or Range or Single or Specific List or Null.
Programmatic Pattern: ((RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | MSAnalyteGroupP) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeScanTime
The duration of time allowed to pass between each spectral acquisition. When ColumnPrimeAcquisitionMode is DataDependent, this value refers to the duration for measuring spectra from the intact ions. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired.
Default Calculation: Set to 0.2 seconds unless a method is given.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null.
Programmatic Pattern: (RangeP[0.015*Second, 10*Second] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeFragmentMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for product ions. When ColumnPrimeAcquisitionMode is DataDependent|DataIndependent, all of the product ions in consideration for measurement. Null if ColumnPrimeFragment is False.
Default Calculation: When ColumnPrimeFragment is False, set to Null. Otherwise, 20 Gram/Mole to the maximum ColumnPrimeMassDetection.
Pattern Description: DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null.
Programmatic Pattern: (({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeCollisionEnergy
The voltage by which intact ions are accelerated through inert gas in order to dissociate them into measurable fragment ion species when ColumnPrimeFragment is True. ColumnPrimeCollisionEnergy cannot be defined simultaneously with ColumnPrimeCollisionEnergyMassProfile.
Default Calculation: Is automatically set to 40 Volt when ColumnPrimeFragment is True, otherwise is set to Null.
Pattern Description: Greater than or equal to 5 volts and less than or equal to 255 volts or greater than or equal to -180 volts and less than or equal to 5 volts or list of one or more greater than or equal to 5 volts and less than or equal to 180 volts or greater than or equal to -180 volts and less than or equal to 5 volts entries or Null.
Programmatic Pattern: (((RangeP[5*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {(RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt])..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeCollisionEnergyMassProfile
The relationship of collision energy with the ColumnPrimeMassDetection.
Default Calculation: Set to ColumnPrimeCollisionEnergyMassScan if defined; otherwise, set to Null.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeCollisionEnergyMassScan
The collision energy profile at the end of the scan from ColumnPrimeCollisionEnergy or ColumnPrimeCollisionEnergyScanProfile, as related to analyte mass.
Pattern Description: Constant or Range or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] | RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt]) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeFragmentScanTime
The duration of the spectral scanning for each fragmentation of an intact ion when ColumnPrimeAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to the same value as ScanTime if ColumnPrimeAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null.
Programmatic Pattern: ((Alternatives[RangeP[0.015*Second, 10*Second]]) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeAcquisitionSurvey
The number of intact ions to consider for fragmentation and product ion measurement in every measurement cycle when ColumnPrimeAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to 10 if ColumnPrimeAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 30 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 30, 1] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeMinimumThreshold
The minimum number of total ions detected within ScanTime durations needed to trigger the start of data dependent acquisition when ColumnPrimeAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to (100000/Second)*ScanTime if ColumnPrimeAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null.
Programmatic Pattern: (RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeAcquisitionLimit
The maximum number of total ions for a specific intact ion when ColumnPrimeAcquisitionMode is set to DataDependent. When this value is exceeded, acquisition will switch to fragmentation of the next candidate ion.
Default Calculation: Automatically inherited from supplied method if ColumnPrimeAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null.
Programmatic Pattern: (RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeCycleTimeLimit
The maximum possible computed duration of all of the scans for the intact and fragmentation measurements when ColumnPrimeAcquisitionMode is set to DataDependent.
Default Calculation: Calculated from the ColumnPrimeAcquisitionSurvey, ColumnPrimeScanTime, and ColumnPrimeFragmentScanTime.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or Null.
Programmatic Pattern: (RangeP[0.015*Second, 20000*Second] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeExclusionDomain
THe time range when the ColumnPrimeExclusionMasses are omitted in the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire ColumnPrimeAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeExclusionMass
The intact ions (Target Mass) to omit. When the Mode is set to All, the mass is excluded for the entire ExclusionDomain. When set to Once, the mass is excluded in the first survey appearance, but considered for consequent ones.
Default Calculation: If any ColumnPrimeExclusionMode-related options are set (e.g. ColumnPrimeExclusionMassTolerance), a target mass of the first Analyte (if not in ColumnPrimeInclusionMasses) is chosen and retention time is set to 0*Minute.
Pattern Description: List of one or more {Mode, Target Mass} entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | {{All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]}..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeExclusionMassTolerance
The range above and below each ion in ColumnPrimeExclusionMasses to consider for omission when ColumnPrimeExclusionMass is set to All or Once.
Default Calculation: If ColumnPrimeExclusionMass -> All or Once, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: (RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeExclusionRetentionTimeTolerance
The range of time above and below the ColumnPrimeExclusionDomain to consider for exclusion.
Default Calculation: If ColumnPrimeExclusionMass and ColumnPrimeExclusionDomain options are set, this is set to 10 seconds; otherwise, Null.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 3600 seconds or Null.
Programmatic Pattern: (RangeP[0*Second, 3600*Second] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeInclusionDomain
The time range when the ColumnPrimeInclusionMass applies with respect to the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire ColumnPrimeAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeInclusionMass
The ions (Target Mass) to prioritize during the survey scan for further fragmentation when ColumnPrimeAcquisitionMode is DataDependent. ColumnPrimeInclusionMass set to Only will solely be considered for surveys. When Mode is Preferential, the InclusionMass will be prioritized for survey.
Default Calculation: When ColumnPrimeInclusionMode Only or Preferential, an entry mass is added based on the mass of the most salient analyte of the sample.
Pattern Description: List of one or more {Mode, Target Mass} entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | {{Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]}..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeInclusionCollisionEnergy
The overriding collision energy value that can be applied to the ColumnPrimeInclusionMass. Null will default to the ColumnPrimeCollisionEnergy option and related.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 volts and less than or equal to 255 volts or list of one or more greater than or equal to 0 volts and less than or equal to 255 volts entries or Null.
Programmatic Pattern: ((RangeP[0*Volt, 255*Volt] | {RangeP[0*Volt, 255*Volt]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeInclusionDeclusteringVoltage
The overriding source voltage value that can be applied to the ColumnPrimeInclusionMass. Null will default to the ColumnPrimeDeclusteringVoltage option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 150*Volt] | {RangeP[0.1*Volt, 150*Volt]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeInclusionChargeState
The maximum charge state of the ColumnPrimeInclusionMass to also consider for inclusion. For example, if this is set to 3 and the polarity is Positive, then +1,+2,+3 charge states will be considered as well.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 entries or Null.
Programmatic Pattern: ((RangeP[0, 6, 1] | {RangeP[0, 6, 1]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeInclusionScanTime
The overriding scan time duration that can be applied to the ColumnPrimeInclusionMass for the consequent fragmentation. Null will default to the ColumnPrimeFragmentScanTime option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {RangeP[0.015*Second, 10*Second]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeInclusionMassTolerance
The range above and below each ion in ColumnPrimeInclusionMass to consider for prioritization. For example, if set to 0.5 Gram/Mole, the total range is 1 Gram/Mole.
Default Calculation: Set to 0.5 Gram/Mole if ColumnPrimeInclusionMass is given; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: ((Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]]) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeSurveyChargeStateExclusion
Indicates if redundant ions that differ by ionic charge (+1/-1, +2/-2, etc.) should be excluded and if ColumnPrimeChargeState exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the ColumnPrimeChargeState options are set; otherwise, False.
Pattern Description: True or False or Null.
Programmatic Pattern: ((Alternatives[BooleanP]) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeSurveyIsotopeExclusion
Indicates if redundant ions that differ by isotopic mass (e.g. 1, 2 Gram/Mole) should be excluded and if ColumnPrimeMassIsotope exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the ColumnPrimeIsotopeExclusion options are set; otherwise, False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeChargeStateExclusionLimit
The number of ions to survey first with exclusion by ionic state. For example, if ColumnPrimeAcquisitionSurvey is 10 and this option is 5, then 5 ions will be surveyed with charge-state exclusion. For candidate ions of rank 6 to 10, no exclusion will be performed.
Default Calculation: Inherited from any supplied method; otherwise, set the same to ColumnPrimeAcquisitionSurvey, if any ChargeState option is set.
Pattern Description: Greater than or equal to 0 and less than or equal to 30 in increments of 1 or Null.
Programmatic Pattern: (RangeP[0, 30, 1] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeChargeStateExclusion
The specific ionic states of intact ions to redundantly exclude from the survey for further fragmentation/acquisition. 1 refers to +1/-1, 2 refers to +2/-2, etc.
Default Calculation: When ColumnPrimeSurveyChargeStateExclusion is True, set to {1,2}; otherwise, Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 entries or Null.
Programmatic Pattern: ((RangeP[1, 6, 1] | {RangeP[1, 6, 1]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeChargeStateMassTolerance
The range of m/z to consider for exclusion by ionic state property when ColumnPrimeSurveyChargeStateExclusion is True.
Default Calculation: When ColumnPrimeSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: (RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeIsotopicExclusion
The m/z difference between monoisotopic ions as a criterion for survey exclusion.
Default Calculation: When ColumnPrimeSurveyIsotopeExclusion is True, set to 1 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole entries or Null.
Programmatic Pattern: (((Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]]) | {(Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]])..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeIsotopeRatioThreshold
The minimum relative magnitude between monoisotopic ions in order to be considered an isotope for exclusion.
Default Calculation: When ColumnPrimeSurveyIsotopeExclusion is True, set to 0.1; otherwise, Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 entries or Null.
Programmatic Pattern: ((RangeP[0, 1] | {RangeP[0, 1]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeIsotopeDetectionMinimum
The acquisition rate of a given intact mass to consider for isotope exclusion in the survey.
Default Calculation: When ColumnPrimeSurveyIsotopeExclusion is True, set to 10 1/Second; otherwise, Null.
Pattern Description: Greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(1/Second)] | {GreaterEqualP[0*(1/Second)]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeIsotopeMassTolerance
The range of m/z around a mass to consider for exclusion. This applies for both ChargeState and mass shifted Isotope. If set to 0.5 Gram/Mole, then the total range should be 1 Gram/Mole.
Default Calculation: When ColumnPrimeSurveyIsotopeExclusion or ColumnPrimeSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: ((Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]]) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeIsotopeRatioTolerance
The range of relative magnitude around ColumnPrimeIsotopeRatio to consider for isotope exclusion.
Default Calculation: If ColumnPrimeSurveyIsotopeExclusion is True, set to 30*Percent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: ((Alternatives[RangeP[0*Percent, 100*Percent]]) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeNeutralLoss
A neutral loss scan is performed on ESI-QQQ mass spectrometry by scanning the sample through the first quadrupole (Q1). The ions are then fragmented in the collision cell. The second mass analyzer is then scanned with a fixed offset to MS1. This option represents the value of this offset.
Default Calculation: Is set to 500 g/mol if using NeutralIonLoss as the ColumnPrimeAcquisitionMode, and is Null in other modes.
Pattern Description: Greater than 0 grams per mole or Null.
Programmatic Pattern: (GreaterP[0*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeDwellTime
The duration of time for which spectra are acquired at the specific mass detection value for SelectedIonMonitoring and MultipleReactionMonitoring mode in ESI-QQQ.
Default Calculation: Is automatically set to 200 microsecond if ColumnPrimeAcquisitionMode is in SelectedIonMonitoring or MultipleReactionMonitoring mode.
Pattern Description: Greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds entries or Null.
Programmatic Pattern: ((RangeP[5*Millisecond, 2000*Millisecond] | {RangeP[5*Millisecond, 2000*Millisecond]..}) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeCollisionCellExitVoltage
Also known as the Collision Cell Exit Potential (CXP). This value focuses and accelerates the ions out of collision cell (Q2) and into 2nd mass analyzer (MS 2). This potential is tuned to ensure successful ion acceleration out of collision cell and into MS2, and can be adjusted to reach the maximal signal intensity. This option is unique to ESI-QQQ for now, and only required when Fragment ->True and/or in ScanMode that achieves tandem mass feature (PrecursorIonScan, NeutralIonLoss,ProductIonScan,MultipleReactionMonitoring). For non-tandem mass ScanMode (FullScan and SelectedIonMonitoring) and other massspectrometer (ESI-QTOF and MALDI-TOF), this option is resolved to Null.
Default Calculation: For TripleQuadrupole as the MassAnalyzer, is set to first CollisionCellExitVoltage, otherwise set to Null.
Pattern Description: Greater than or equal to -55 volts and less than or equal to 55 volts or Null.
Programmatic Pattern: (RangeP[-55*Volt, 55*Volt] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeMassDetectionStepSize
Indicate the step size for mass collection in range when using TripleQuadruploe as the MassAnalyzer.
Default Calculation: Is set to first CollisionCellExitVoltage
Pattern Description: Greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or Null.
Programmatic Pattern: (RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeMultipleReactionMonitoringAssays
In ESI-QQQ mass spectrometry analysis, the ion corresponding to the compound of interest is targeted with subsequent fragmentation of that target ion to produce a range of daughter ions. One (or more) of these fragment daughter ions can be selected for quantitation purposes. Only compounds that meet both these criteria, i.e. specific parent ion and specific daughter ions corresponding to the mass of the molecule of interest are detected within the mass spectrometer. The mass assays (MS1/MS2 mass value combinations) for each scan, along with the CollisionEnergy and DwellTime (length of time of each scan).
Default Calculation: Is set based on ColumnPrimeMassDetection, ColumnPrimeCollisionEnergy, ColumnPrimeDwellTime and ColumnPrimeFragmentMassDetection.
Pattern Description: Individual Multiple Reaction Monitoring Assay or {Multiple Reaction Monitoring Assays} or None.
Programmatic Pattern: (({{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..} | {{{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..}} | (Null | {Null})) | Automatic) | Null
Index Matches to: ColumnPrimeAcquisitionWindow
ColumnPrimeAbsorbanceWavelength
The wavelength of light passed through the flow for the UVVis and Detector for detection during column prime. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 500*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 490*Nanometer, 1*Nanometer] ;; RangeP[200*Nanometer, 500*Nanometer, 1*Nanometer]) | Automatic) | Null
ColumnPrimeWavelengthResolution
The increment in wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector during column prime.
Default Calculation: Automatically set to the same as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[1.2*Nanometer, 12.*Nanometer] | Automatic) | Null
ColumnPrimeUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the sample for the PhotoDiodeArray (PDA) detector for ColumnPrime measurements.
Default Calculation: Automatically set to the same as the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
ColumnPrimeAbsorbanceSamplingRate
The number of times an absorbance measurement is made per second during column prime. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second), 1*(1/Second)] | Automatic) | Null
Column Flush
ColumnFlushTemperature
The column's temperature at which the column flush gradient is run. If ColumnFlushTemperature is set to Ambient, column oven temperature control is not used. Otherwise, ColumnFlushTemperature is maintained by temperature control of the column oven.
Default Calculation: Automatically set to the corresponding gradient temperature specified in the ColumnFlushGradient option or the column temperature for the column flush in the InjectionTable option; otherwise, set as the first value of the ColumnTemperature option.
Pattern Description: Ambient or greater than or equal to 30 degrees Celsius and less than or equal to 90 degrees Celsius or Null.
Programmatic Pattern: ((RangeP[30*Celsius, 90*Celsius] | Ambient) | Automatic) | Null
ColumnFlushGradientA
The composition of BufferA within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientA->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferA in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientB, ColumnFlushGradientC, and ColumnFlushGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer A Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushGradientB
The composition of BufferB within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientB->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferB in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientA, ColumnFlushGradientC, and ColumnFlushGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer B Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushGradientC
The composition of BufferC within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientC->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferC in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientA, ColumnFlushGradientB, and ColumnFlushGradientD options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer C Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushGradientD
The composition of BufferD within the flow, defined for specific time points for column flush. The composition is linearly interpolated for the intervening periods between the defined time points. For example for ColumnFlushGradientD->{{0 Minute, 10 Percent},{30 Minute, 90 Percent}}, the percentage of BufferD in the flow will rise such that at 15 minutes, the composition should be 50 Percent.
Default Calculation: If ColumnFlushGradient option is specified, set from it or implicitly determined from the ColumnFlushGradientA, ColumnFlushGradientB, and ColumnFlushGradientC options such that the total amounts to 100%.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or list of one or more {Time, Buffer D Composition} entries or Null.
Programmatic Pattern: ((RangeP[0*Percent, 100*Percent] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent]}..}) | Automatic) | Null
ColumnFlushFlowRate
The net speed of the fluid flowing through the pump inclusive of the composition of BufferA, BufferB, BufferC, and BufferD specified in the ColumnFlushGradient options during column flush. This speed is linearly interpolated such that consecutive entries of {Time, Flow Rate} will define the intervening fluid speed. For example, {{0 Minute, 0.3 Milliliter/Minute},{30 Minute, 0.5 Milliliter/Minute}} means flow rate of 0.4 Milliliter/Minute at 15 minutes into the run.
Default Calculation: If ColumnFlushGradient option is specified, automatically set from the method given in the ColumnFlushGradient option. If NominalFlowRate of the column model is specified, set to lesser of the NominalFlowRate for each of the columns, guard columns or the instrument's MaxFlowRate. Otherwise set to 1 Milliliter / Minute.
Pattern Description: Greater than or equal to 0 milliliters per minute and less than or equal to 2 milliliters per minute or list of one or more {Time, Flow Rate} entries or Null.
Programmatic Pattern: ((RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnFlushGradient
The composition of different specified buffers in BufferA, BufferB, BufferC and BufferD over time in the fluid flow for column prime. Specific parameters of a gradient object can be overridden by specific options.
Default Calculation: Automatically set to best meet the values specified in ColumnFlushGradient options (e.g. ColumnFlushGradientA, ColumnFlushGradientB, ColumnFlushGradientC, ColumnFlushGradientD, ColumnFlushFlowRate).
Pattern Description: An object of type or subtype Object[Method, Gradient] or list of one or more {Time, Buffer A Composition, Buffer B Composition, Buffer C Composition, Buffer D Composition, Flow Rate} entries or Null.
Programmatic Pattern: ((ObjectP[Object[Method, Gradient]] | {{RangeP[0*Minute, $MaxExperimentTime], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*Percent, 100*Percent], RangeP[0*(Milliliter/Minute), 2*(Milliliter/Minute)]}..}) | Automatic) | Null
ColumnFlushIonMode
Indicates if positively or negatively charged ions are analyzed.
Default Calculation: Set to the first IonMode for an analyte input sample.
Pattern Description: Negative or Positive or Null.
Programmatic Pattern: (IonModeP | Automatic) | Null
ColumnFlushMassSpectrometryMethod
The previously specified instruction(s) for the analyte ionization, selection, fragmentation, and detection.
Default Calculation: If ColumnFlush samples exist and MassSpectrometryMethod is specified, then set to the first available ColumnFlushMassSpectrometryMethod.
Pattern Description: An object of type or subtype Object[Method, MassAcquisition] or New or Null.
Programmatic Pattern: ((ObjectP[Object[Method, MassAcquisition]] | New) | Automatic) | Null
ColumnFlushESICapillaryVoltage
The absolute voltage applied to the tip of the stainless steel capillary tubing in order to produce charged droplets. Adjust this voltage to maximize sensitivity. Most compounds are optimized between 0.5 and 3.2 kV in ESI positive ion mode and 0.5 and 2.6 in ESI negative ion mode, but can be altered according to sample type. For low flow applications, best sensitivity will be achieved with a relatively high value in ESI positive (e.g. 3.0 kV), for columnFlush flow UPLC a value of 0.5 kV is typically best for maximum sensitivity.
Default Calculation: Is automatically set to the first ESICapillaryVoltage.
Pattern Description: Greater than or equal to -4 kilovolts and less than or equal to 5 kilovolts or Null.
Programmatic Pattern: (RangeP[-4*Kilovolt, 5*Kilovolt] | Automatic) | Null
ColumnFlushDeclusteringVoltage
The voltage offset between the ion block (the reduced pressure chamber of the source block) and the stepwave ion guide (the optics before the quadrupole mass analyzer). This voltage attracts charged ions in the spray being produced from the capillary tip into the ion block leading into the mass spectrometer. This voltage is typically set to 25-100 V and its tuning has little effect on sensitivity compared to other options (e.g. ColumnFlushStepwaveVoltage).
Default Calculation: Is automatically set to any specified MassAcquisition method; otherwise, set to 40 Volt.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 150*Volt] | Automatic) | Null
ColumnFlushStepwaveVoltage
The voltage offset between the 1st and 2nd stage of the stepwave ion guide which leads ions coming from the sample cone towards the quadrupole mass analyzer. This voltage normally optimizes between 25 and 150 V and should be adjusted for sensitivity depending on compound and charge state. For multiply charged species it is typically set to to 40-50 V, and higher for singly charged species. In general higher cone voltages (120-150 V) are needed for larger mass ions such as intact proteins and monoclonal antibodies. It also has greatest effect on in-source fragmentation and should be decreased if in-source fragmentation is observed but not desired.
Default Calculation: Is automatically set to the first StepwaveVoltage.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 200 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 200*Volt] | Automatic) | Null
ColumnFlushSourceTemperature
The temperature setting of the source block. Heating the source block discourages condensation and decreases solvent clustering in the reduced vacuum region of the source. This temperature setting is flow rate and sample dependent. Typical values are between 60 to 120 Celsius. For thermally labile analytes, a lower temperature setting is recommended.
Default Calculation: Is automatically set to the first SourceTemperature.
Pattern Description: Greater than or equal to 25 degrees Celsius and less than or equal to 150 degrees Celsius or Null.
Programmatic Pattern: (RangeP[25*Celsius, 150*Celsius] | Automatic) | Null
ColumnFlushDesolvationTemperature
The temperature setting for the ESI desolvation heater that controls the nitrogen gas temperature used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to ColumnFlushDesolvationGasFlow, this setting is dependent on solvent flow rate and composition. A typical range is from 150 to 650 Celsius.
Default Calculation: Is automatically set to the first DesolvationTemperature.
Pattern Description: Greater than or equal to 20 degrees Celsius and less than or equal to 650 degrees Celsius or Null.
Programmatic Pattern: (RangeP[20*Celsius, 650*Celsius] | Automatic) | Null
ColumnFlushDesolvationGasFlow
The rate at which nitrogen gas is flowed around the ESI capillary. It is used for solvent evaporation to produce single gas phase ions from the ion spray. Similar to ColumnFlushDesolvationTemperature, this setting is dependent on solvent flow rate and composition. Higher desolvation temperatures usually result in increased sensitivity, but too high values can cause spray instability. Typical values are between 300 to 1200 L/h.
Default Calculation: Is automatically set to the first DesolvationGasFlow.
Pattern Description: Greater than or equal to 55 liters per hour and less than or equal to 1200 liters per hour or greater than or equal to 0 pounds
‐force per inch squared and less than or equal to 85 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[55*(Liter/Hour), 1200*(Liter/Hour)] | RangeP[0*PSI, 85*PSI]) | Automatic) | Null
ColumnFlushConeGasFlow
The rate at which nitrogen gas is flowed around the sample inlet cone (the spherical metal plate acting as a first gate between the sprayer and the reduced pressure chamber, the ion block). This gas flow is used to minimize the formation of solvent ion clusters. It also helps reduce adduct ions and directing the spray into the ion block while keeping the sample cone clean. Typical values are between 0 and 150 L/h.
Default Calculation: Is automatically set to the first ConeGasFlow.
Pattern Description: Greater than or equal to 0 liters per hour and less than or equal to 300 liters per hour or greater than or equal to 30 pounds
‐force per inch squared and less than or equal to 50 pounds
‐force per inch squared or Null.
Programmatic Pattern: ((RangeP[0*(Liter/Hour), 300*(Liter/Hour)] | RangeP[30*PSI, 50*PSI]) | Automatic) | Null
ColumnFlushAcquisitionWindow
The time range with respect to the chromatographic separation to conduct analyte ionization, selection/survey, optional fragmentation, and detection.
Default Calculation: Set to the entire gradient window 0 Minute to the last time point in ColumnFlushGradient.
Pattern Description: A span from anything greater than or equal to 0 minutes and less than or equal to 8 hours to anything greater than or equal to 0 minutes and less than or equal to 8 hours or Null.
Programmatic Pattern: (RangeP[0*Minute, 8*Hour] ;; RangeP[0*Minute, 8*Hour] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushAcquisitionMode
The method by which spectra are collected. DataDependent will depend on the properties of the measured mass spectrum of the intact ions. DataIndependent will systemically scan through all of the intact ions. MS1FullScan will focus on defined intact masses. MS1MS2 will focus on fragmented masses.
Default Calculation: Set to MS1FullScan unless DataDependent related options are set, then set to DataDependent.
Pattern Description: DataIndependent, DataDependent, MS1FullScan, MS1MS2ProductIonScan, SelectedIonMonitoring, NeutralIonLoss, PrecursorIonScan, or MultipleReactionMonitoring or Null.
Programmatic Pattern: (MSAcquisitionModeP | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushFragment
Indicates if ions should be collided with neutral gas and dissociated in order to measure the resulting product ions. Also known as tandem mass spectrometry or MS/MS (as opposed to MS).
Default Calculation: Set to True if ColumnFlushAcquisitionMode is MS1MS2ProductIonScan, DataDependent, or DataIndependent. Set True if any of the Fragmentation related options are set (e.g. ColumnFlushFragmentMassDetection).
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for intact masses. When ColumnFlushFragment is True, the intact ions will be selected for fragmentation.
Default Calculation: For ColumnFlushFragment -> False, automatically set to one of three default mass ranges according to the molecular weight of the ColumnFlushAnalytes to encompass them.
Pattern Description: All or Range or Single or Specific List or Null.
Programmatic Pattern: ((RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | {RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]..} | RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] ;; RangeP[2*(Gram/Mole), 100000*(Gram/Mole)] | MSAnalyteGroupP) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushScanTime
The duration of time allowed to pass between each spectral acquisition. When ColumnFlushAcquisitionMode is DataDependent, this value refers to the duration for measuring spectra from the intact ions. Increasing this value improves sensitivity whereas decreasing this value allows for more data points and spectra to be acquired.
Default Calculation: Set to 0.2 seconds unless a method is given.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null.
Programmatic Pattern: (RangeP[0.015*Second, 10*Second] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushFragmentMassDetection
The lowest and the highest mass-to-charge ratio (m/z) to be recorded or selected for product ions. When ColumnFlushAcquisitionMode is DataDependent|DataIndependent, all of the product ions in consideration for measurement. Null if ColumnFlushFragment is False.
Default Calculation: When ColumnFlushFragment is False, set to Null. Otherwise, 20 Gram/Mole to the maximum ColumnFlushMassDetection.
Pattern Description: DataDependent or DataIndependent or MultipleReactionMonitoring or PrecursorIonScan or ProductIonScan or Null.
Programmatic Pattern: (({RangeP[5*(Gram/Mole), 16000*(Gram/Mole)]..} | RangeP[5*(Gram/Mole), 16000*(Gram/Mole)] ;; RangeP[100*(Gram/Mole), 16000*(Gram/Mole)] | All | Null | {RangeP[5*(Gram/Mole), 2000*(Gram/Mole)]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushCollisionEnergy
The voltage by which intact ions are accelerated through inert gas in order to dissociate them into measurable fragment ion species when ColumnFlushFragment is True. Cannot be defined simultaneously with ColumnFlushCollisionEnergyMassProfile.
Default Calculation: Is automatically set to 40 Volt when ColumnFlushFragment is True, otherwise is set to Null.
Pattern Description: Greater than or equal to 5 volts and less than or equal to 255 volts or greater than or equal to -180 volts and less than or equal to 5 volts or list of one or more greater than or equal to 5 volts and less than or equal to 180 volts or greater than or equal to -180 volts and less than or equal to 5 volts entries or Null.
Programmatic Pattern: (((RangeP[5*Volt, 255*Volt] | RangeP[-180*Volt, 5*Volt]) | {(RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt])..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushCollisionEnergyMassProfile
The relationship of collision energy with the ColumnFlushMassDetection.
Default Calculation: Set to ColumnFlushCollisionEnergyMassScan if defined; otherwise, set to Null.
Pattern Description: A span from anything greater than or equal to 0.1 volts and less than or equal to 255 volts to anything greater than or equal to 0.1 volts and less than or equal to 255 volts or Null.
Programmatic Pattern: (RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushCollisionEnergyMassScan
The collision energy profile at the end of the scan from ColumnFlushCollisionEnergy or ColumnFlushCollisionEnergyScanProfile, as related to analyte mass.
Pattern Description: Constant or Range or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 255*Volt] | RangeP[0.1*Volt, 255*Volt] ;; RangeP[0.1*Volt, 255*Volt]) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushFragmentScanTime
The duration of the spectral scanning for each fragmentation of an intact ion when ColumnFlushAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to the same value as ScanTime if ColumnFlushAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or Null.
Programmatic Pattern: ((Alternatives[RangeP[0.015*Second, 10*Second]]) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushAcquisitionSurvey
The number of intact ions to consider for fragmentation and product ion measurement in every measurement cycle when ColumnFlushAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to 10 if ColumnFlushAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 30 in increments of 1 or Null.
Programmatic Pattern: (RangeP[1, 30, 1] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushMinimumThreshold
The minimum number of total ions detected within ScanTime durations needed to trigger the start of data dependent acquisition when ColumnFlushAcquisitionMode is set to DataDependent.
Default Calculation: Automatically set to (100000/Second)*ScanTime if ColumnFlushAcquisitionMode is DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null.
Programmatic Pattern: (RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushAcquisitionLimit
The maximum number of total ions for a specific intact ion when ColumnFlushAcquisitionMode is set to DataDependent. When this value is exceeded, acquisition will switch to fragmentation of the next candidate ion.
Default Calculation: Automatically inherited from supplied method if ColumnFlushAcquisitionMode is set to DataDependent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 ArbitraryUnits and less than or equal to 8000000 ArbitraryUnits or Null.
Programmatic Pattern: (RangeP[0*ArbitraryUnit, 8000000*ArbitraryUnit] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushCycleTimeLimit
The maximum possible computed duration of all of the scans for the intact and fragmentation measurements when ColumnFlushAcquisitionMode is set to DataDependent.
Default Calculation: Calculated from the ColumnFlushAcquisitionSurvey, ColumnFlushScanTime, and ColumnFlushFragmentScanTime.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 20000 seconds or Null.
Programmatic Pattern: (RangeP[0.015*Second, 20000*Second] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushExclusionDomain
The tune range when the ColumnFlushExclusionMasses are omitted in the chromatogram. Full indicates for the entire period.
Default Calculation: Set to the entire ColumnFlushAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushExclusionMass
The intact ions (Target Mass) to omit. When the Mode is set to All, the mass is excluded for the entire ExclusionDomain. When the Mode is set to Once, the Mass is excluded in the first survey appearance, but considered for consequent ones.
Default Calculation: If any ColumnFlushExclusionMode-related options are set (e.g. ColumnFlushExclusionMassTolerance), a target mass of the first Analyte (if not in ColumnFlushInclusionMasses) is chosen and retention time is set to 0*Minute.
Pattern Description: List of one or more {Mode, Target Mass} entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]} | {{All | Once, RangeP[2*(Gram/Mole), 100000*(Gram/Mole)]}..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushExclusionMassTolerance
The range above and below each ion in ColumnFlushExclusionMasses to consider for omission when ColumnFlushExclusionMass is set to All or Once.
Default Calculation: If ColumnFlushExclusionMass -> All or Once, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: (RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushExclusionRetentionTimeTolerance
The range of time above and below the ColumnFlushExclusionDomain to consider for exclusion.
Default Calculation: If ColumnFlushExclusionMass and ColumnFlushExclusionDomain options are set, this is set to 10 seconds; otherwise, Null.
Pattern Description: Greater than or equal to 0 seconds and less than or equal to 3600 seconds or Null.
Programmatic Pattern: (RangeP[0*Second, 3600*Second] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushInclusionDomain
The time range when the ColumnFlushInclusionMass applies with respect to the chromatogram.
Default Calculation: Set to the entire ColumnFlushAcquisitionWindow.
Pattern Description: A span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full or list of one or more a span from anything greater than or equal to 0 minutes to anything greater than or equal to 0 minutes or Full entries or Null.
Programmatic Pattern: (((GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full) | {(GreaterEqualP[0*Minute] ;; GreaterEqualP[0*Minute] | Full)..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushInclusionMass
The ions (Target Mass) to prioritize during the survey scan for further fragmentation When ColumnFlushAcquisitionMode is DataDependent. ColumnFlushInclusionMass set to Only will solely be considered for surveys. When Mode is Preferential, the InclusionMass will be prioritized for survey.
Default Calculation: When ColumnFlushInclusionMode Only or Preferential, an entry mass is added based on the mass of the most salient analyte of the sample.
Pattern Description: List of one or more {Mode, Target Mass} entries or {Mode, Target Mass} or Null.
Programmatic Pattern: (({Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]} | {{Only | Preferential, RangeP[2*(Gram/Mole), 4000*(Gram/Mole)]}..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushInclusionCollisionEnergy
The overriding collision energy value that can be applied to the ColumnFlushInclusionMass. Null will default to the ColumnFlushCollisionEnergy option and related.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 volts and less than or equal to 255 volts or list of one or more greater than or equal to 0 volts and less than or equal to 255 volts entries or Null.
Programmatic Pattern: ((RangeP[0*Volt, 255*Volt] | {RangeP[0*Volt, 255*Volt]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushInclusionDeclusteringVoltage
The overriding source voltage value that can be applied to the ColumnFlushInclusionMass. Null will default to the ColumnFlushDeclusteringVoltage option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.1 volts and less than or equal to 150 volts or list of one or more greater than or equal to 0.1 volts and less than or equal to 150 volts entries or Null.
Programmatic Pattern: ((RangeP[0.1*Volt, 150*Volt] | {RangeP[0.1*Volt, 150*Volt]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushInclusionChargeState
The maximum charge state of the ColumnFlushInclusionMass to also consider for inclusion. For example, if this is set to 3 and the polarity is Positive, then +1,+2,+3 charge states will be considered as well.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 0 and less than or equal to 6 in increments of 1 entries or Null.
Programmatic Pattern: ((RangeP[0, 6, 1] | {RangeP[0, 6, 1]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushInclusionScanTime
The overriding scan time duration that can be applied to the ColumnFlushInclusionMass for the consequent fragmentation. Null will default to the ColumnFlushFragmentScanTime option.
Default Calculation: Inherited from any supplied method.
Pattern Description: Greater than or equal to 0.015 seconds and less than or equal to 10 seconds or list of one or more greater than or equal to 0.015 seconds and less than or equal to 10 seconds entries or Null.
Programmatic Pattern: ((RangeP[0.015*Second, 10*Second] | {RangeP[0.015*Second, 10*Second]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushInclusionMassTolerance
The range above and below each ion in ColumnFlushInclusionMass to consider for prioritization. For example, if set to 0.5 Gram/Mole, the total range is 1 Gram/Mole.
Default Calculation: Set to 0.5 Gram/Mole if ColumnFlushInclusionMass is given; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: ((Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]]) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushSurveyChargeStateExclusion
Indicates if redundant ions that differ by ionic charge (+1/-1, +2/-2, etc.) should be excluded and if ColumnFlushChargeState exclusion-related options should be automatically filled in.
Default Calculation: Set to True, if any of the ColumnFlushChargeState options are set; otherwise, False.
Pattern Description: True or False or Null.
Programmatic Pattern: ((Alternatives[BooleanP]) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushSurveyIsotopeExclusion
Indicates if redundant ions that differ by isotopic mass (e.g. 1, 2 Gram/Mole) should be excluded and if ColumnFlushMassIsotope exclusion-related options should be automatically filled.
Default Calculation: Set to True, if any of the ColumnFlushIsotopeExclusion options are set; otherwise, False.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushChargeStateExclusionLimit
The number of ions to survey first with exclusion by ionic state. For example, if ColumnFlushAcquisitionSurvey is 10 and this option is 5, then 5 ions will be surveyed with charge-state exclusion. For candidate ions of rank 6 to 10, no exclusion will be performed.
Default Calculation: Inherited from any supplied method; otherwise, set the same to ColumnFlushAcquisitionSurvey, if any ChargeState option is set.
Pattern Description: Greater than or equal to 0 and less than or equal to 30 in increments of 1 or Null.
Programmatic Pattern: (RangeP[0, 30, 1] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushChargeStateExclusion
The specific ionic states of intact ions to redundantly exclude from the survey for further fragmentation/acquisition. 1 refers to +1/-1, 2 refers to +2/-2, etc.
Default Calculation: When ColumnFlushSurveyChargeStateExclusion is True, set to {1,2}; otherwise, Null.
Pattern Description: Greater than or equal to 1 and less than or equal to 6 in increments of 1 or list of one or more greater than or equal to 1 and less than or equal to 6 in increments of 1 entries or Null.
Programmatic Pattern: ((RangeP[1, 6, 1] | {RangeP[1, 6, 1]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushChargeStateMassTolerance
The range of m/z to consider for exclusion by ionic state property when ColumnFlushSurveyChargeStateExclusion is True.
Default Calculation: When ColumnFlushSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: (RangeP[0*(Gram/Mole), 3000*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushIsotopicExclusion
The m/z difference between monoisotopic ions as a criterion for survey exclusion.
Default Calculation: When ColumnFlushSurveyIsotopeExclusion is True, set to 1 Gram/Mole; otherwise, Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or list of one or more greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole entries or Null.
Programmatic Pattern: (((Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]]) | {(Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]])..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushIsotopeRatioThreshold
The minimum relative magnitude between monoisotopic ions in order to be considered an isotope for exclusion.
Default Calculation: When ColumnFlushSurveyIsotopeExclusion is True, set to 0.1; otherwise, Null.
Pattern Description: Greater than or equal to 0 and less than or equal to 1 or list of one or more greater than or equal to 0 and less than or equal to 1 entries or Null.
Programmatic Pattern: ((RangeP[0, 1] | {RangeP[0, 1]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushIsotopeDetectionMinimum
The acquisition rate of a given intact mass to consider for isotope exclusion in the survey.
Default Calculation: When ColumnFlushSurveyIsotopeExclusion is True, set to 10 1/Second; otherwise, Null.
Pattern Description: Greater than or equal to 0 reciprocal seconds or list of one or more greater than or equal to 0 reciprocal seconds entries or Null.
Programmatic Pattern: ((GreaterEqualP[0*(1/Second)] | {GreaterEqualP[0*(1/Second)]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushIsotopeMassTolerance
The range of m/z around a mass to consider for exclusion. This applies for both ChargeState and mass shifted Isotope. If set to 0.5 Gram/Mole, then the total range should be 1 Gram/Mole.
Default Calculation: When ColumnFlushSurveyIsotopeExclusion or ColumnFlushSurveyChargeStateExclusion is True, set to 0.5 Gram/Mole; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 grams per mole and less than or equal to 3000 grams per mole or Null.
Programmatic Pattern: ((Alternatives[RangeP[0*(Gram/Mole), 3000*(Gram/Mole)]]) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushIsotopeRatioTolerance
The range of relative magnitude around ColumnFlushIsotopeRatio to consider for isotope exclusion.
Default Calculation: If ColumnFlushSurveyIsotopeExclusion is True, set to 30*Percent; otherwise, set to Null.
Pattern Description: Greater than or equal to 0 percent and less than or equal to 100 percent or Null.
Programmatic Pattern: ((Alternatives[RangeP[0*Percent, 100*Percent]]) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushNeutralLoss
A neutral loss scan is performed on ESI-QQQ mass spectrometry by scanning the sample through the first quadrupole (Q1). The ions are then fragmented in the collision cell. The second mass analyzer is then scanned with a fixed offset to MS1. This option represents the value of this offset.
Default Calculation: Is set to 500 g/mol if using NeutralIonLoss as the ColumnFlushAcquisitionMode, and is Null in other modes.
Pattern Description: Greater than 0 grams per mole or Null.
Programmatic Pattern: (GreaterP[0*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushDwellTime
The duration of time for which spectra are acquired at the specific mass detection value for SelectedIonMonitoring and MultipleReactionMonitoring mode in ESI-QQQ.
Default Calculation: Is automatically set to 200 microsecond if ColumnFlushAcquisitionMode is in SelectedIonMonitoring or MultipleReactionMonitoring mode.
Pattern Description: Greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds or list of one or more greater than or equal to 5 milliseconds and less than or equal to 2000 milliseconds entries or Null.
Programmatic Pattern: ((RangeP[5*Millisecond, 2000*Millisecond] | {RangeP[5*Millisecond, 2000*Millisecond]..}) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushCollisionCellExitVoltage
Also known as the Collision Cell Exit Potential (CXP). This value focuses and accelerates the ions out of collision cell (Q2) and into 2nd mass analyzer (MS 2). This potential is tuned to ensure successful ion acceleration out of collision cell and into MS2, and can be adjusted to reach the maximal signal intensity. This option is unique to ESI-QQQ for now, and only required when Fragment ->True and/or in ScanMode that achieves tandem mass feature (PrecursorIonScan, NeutralIonLoss,ProductIonScan,MultipleReactionMonitoring). For non-tandem mass ScanMode (FullScan and SelectedIonMonitoring) and other massspectrometer (ESI-QTOF and MALDI-TOF), this option is resolved to Null.
Default Calculation: For TripleQuadrupole as the MassAnalyzer, is set to first CollisionCellExitVoltage, otherwise set to Null.
Pattern Description: Greater than or equal to -55 volts and less than or equal to 55 volts or Null.
Programmatic Pattern: (RangeP[-55*Volt, 55*Volt] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushMassDetectionStepSize
Indicate the step size for mass collection in range when using TripleQuadruploe as the MassAnalyzer.
Default Calculation: Is set to first CollisionCellExitVoltage
Pattern Description: Greater than or equal to 0.01 grams per mole and less than or equal to 1 gram per mole or Null.
Programmatic Pattern: (RangeP[0.01*(Gram/Mole), 1*(Gram/Mole)] | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushMultipleReactionMonitoringAssays
In ESI-QQQ mass spectrometry analysis, the ion corresponding to the compound of interest is targeted with subsequent fragmentation of that target ion to produce a range of daughter ions. One (or more) of these fragment daughter ions can be selected for quantitation purposes. Only compounds that meet both these criteria, i.e. specific parent ion and specific daughter ions corresponding to the mass of the molecule of interest are detected within the mass spectrometer. The mass assays (MS1/MS2 mass value combinations) for each scan, along with the CollisionEnergy and DwellTime (length of time of each scan).
Default Calculation: Is set based on ColumnFlushMassDetection, ColumnFlushCollisionEnergy, ColumnFlushDwellTime and ColumnFlushFragmentMassDetection.
Pattern Description: Individual Multiple Reaction Monitoring Assay or {Multiple Reaction Monitoring Assays} or None.
Programmatic Pattern: (({{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..} | {{{GreaterP[0*(Gram/Mole)], (RangeP[5*Volt, 180*Volt] | RangeP[-180*Volt, 5*Volt]) | Automatic, GreaterP[0*(Gram/Mole)], GreaterP[0*Second] | Automatic}..}} | (Null | {Null})) | Automatic) | Null
Index Matches to: ColumnFlushAcquisitionWindow
ColumnFlushAbsorbanceWavelength
The wavelength of light passed through the flow for the UVVis and Detector for detection during column flush. For PhotoDiodeArray Detector, a 3D data is generated from a spectrum of light passing through the flow cell. Absorbance wavelength in that case represents the wavelength at which a 2D data slice is generated from the 3D data.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceWavelength.
Pattern Description: All or Range or Single or Null.
Programmatic Pattern: ((RangeP[190*Nanometer, 500*Nanometer, 1*Nanometer] | All | RangeP[190*Nanometer, 490*Nanometer, 1*Nanometer] ;; RangeP[200*Nanometer, 500*Nanometer, 1*Nanometer]) | Automatic) | Null
ColumnFlushWavelengthResolution
The increment of wavelength for the range of wavelength of light passed through the flow for absorbance measurement for the instruments with PhotoDiodeArray Detector during column flush.
Default Calculation: Automatically set to the same as the first entry in WavelengthResolution.
Pattern Description: Greater than or equal to 1.2 nanometers and less than or equal to 12. nanometers or Null.
Programmatic Pattern: (RangeP[1.2*Nanometer, 12.*Nanometer] | Automatic) | Null
ColumnFlushUVFilter
Indicates if UV wavelengths (less than 210 nm) should be blocked from being transmitted through the flow for PhotoDiodeArray Detector during column flush.
Default Calculation: Automatically set to the same as the first entry in UVFilter.
Pattern Description: True or False or Null.
Programmatic Pattern: (BooleanP | Automatic) | Null
ColumnFlushAbsorbanceSamplingRate
The number of times an absorbance measurement is made per second during column flush. Lower values will be less susceptible to noise but will record less frequently across time.
Default Calculation: Automatically set to the same as the first entry in AbsorbanceSamplingRate.
Pattern Description: Greater than or equal to 1 reciprocal second and less than or equal to 80 reciprocal seconds in increments of 1 reciprocal second or Null.
Programmatic Pattern: (RangeP[1*(1/Second), 80*(1/Second), 1*(1/Second)] | Automatic) | Null
Model Input
PreparedModelAmount
Indicates the amount of a Model[Sample] specified as input to the experiment function that will be prepared in the PreparedModelContainer. When set to All and the input model sample is not preparable, the entire amount of the input model sample that comes from one of the Products is prepared. The selected product must have both Amount and DefaultContainerModel populated, and it must not be a KitProduct. When set to All and the input model is preparable such as water, 1 Milliliter of the input model sample is prepared.
Default Calculation: Automatically set to 1 Milliliter.
Pattern Description: All or Count or Count or Mass or Volume or Null.
Programmatic Pattern: ((Null | (RangeP[1*Microliter, 20*Liter] | RangeP[1*Milligram, 20*Kilogram] | GreaterP[0*Unit, 1*Unit] | GreaterP[0., 1.] | All)) | Automatic) | Null
Index Matches to: experiment samples
PreparedModelContainer
Indicates the container in which a Model[Sample] specified as input to the experiment function will be prepared.
Default Calculation: If PreparedModelAmount is set to All and the input model has a product associated with both Amount and DefaultContainerModel populated, automatically set to the DefaultContainerModel value in the product. Otherwise, automatically set to Model[Container, Plate, "96-well 2mL Deep Well Plate"].
Pattern Description: An object of type or subtype Model[Container] or Null.
Programmatic Pattern: ((Null | ObjectP[Model[Container]]) | Automatic) | Null
Index Matches to: experiment samples